Abstract
FIB-SEM tomography is a powerful technique that integrates a focused ion beam (FIB) and a scanning electron microscope (SEM) to capture high-resolution imaging data of nanostructures. This approach involves collecting in-plane SEM images and using FIB to remove material layers for imaging subsequent planes, thereby producing image stacks. However, these image stacks in FIB-SEM tomography are subject to the shine-through effect, which makes structures visible from the posterior regions of the current plane. This artifact introduces an ambiguity between image intensity and structures in the current plane, making conventional segmentation methods such as thresholding or the k-means algorithm insufficient. In this study, we propose a multimodal machine learning approach that combines intensity information obtained at different electron beam accelerating voltages to improve the three-dimensional (3D) reconstruction of nanostructures. By treating the increased shine-through effect at higher accelerating voltages as a form of additional information, the proposed method significantly improves segmentation accuracy and leads to more precise 3D reconstructions for real FIB tomography data.
Highlights
-
1.
FIB-SEM tomography can be used to acquire high-resolution image stacks of nanostructures.
-
2.
Machine learning reduces artifacts and ambiguities introduced in FIB-SEM tomography because of the shine-through effect.
-
3.
Our approach treats the shine-through effect as a valuable additional information for precise reconstruction.
-
4.
Using multi-voltage images, our multimodal ML approach improves 3D reconstruction accuracy.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Structural analysis is essential in materials science, enabling the precise characterization and modeling of complex materials. This knowledge facilitates the implementation of these materials in the mass production industry and everyday life [1]. However, high-resolution imaging techniques are typically required to investigate materials with nanoscale features, which limit the range of suitable methods. For instance, when examining hierarchical nanoporous materials such as those in our case with ligament sizes larger than 100 nm and nanopores smaller than 20 nm, conventional techniques, such as X-ray nanotomography, may lack the necessary resolution [2].
Electron microscopy, particularly focused ion beam (FIB) tomography in conjunction with scanning electron microscopy (SEM), offers the essential resolution for analyzing conductive hierarchical nanoporous materials. FIB removes nanometer-thin layers of material after each SEM imaging step, building a stack of images for subsequent three-dimensional (3D) reconstruction. Although FIB-SEM tomography has been applied to the study of nanoporous gold and hierarchical nanoporous gold (HNPG) in various studies [3,4,5], several challenges persist.
First, the variation in slice thickness in FIB tomography images can introduce inaccuracies in 3D reconstructions. A slice repositioning method was proposed to address this issue, and a machine learning (ML) method was adopted to solve this problem from the perspective of an image inpainting problem [6].
The shine-through effect is another challenge, where structures from posterior regions become visible through pores in the currently milled plane [7]. This effect complicates the mapping between actual structure and image intensity [8, 9]. While classical machine learning methods like random forests or k-means clustering are effective in cases where the shine-through effect is minimal [8, 10], the utility of more advanced machine learning techniques for suppressing this effect in cases where it is non-negligible was demonstrated [11, 12]. They used simulated data to train machine learning models and successfully applied these optimized models to real nanostructures. This technique, known as transfer learning, has exhibited notable performance in materials science applications [13].
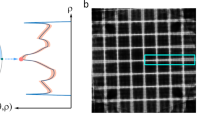
The extent of the shine-through effect is directly related to the beam energy (Fig. 1), allowing its use as an auxiliary method to establish an overdetermined system and collect a comprehensive SEM dataset. In other words, by analyzing images acquired at different voltages, it is possible to separate the shine-through effect signal from the surface area signal, ultimately enabling the reconstruction of hierarchical nanoporous structures with significantly improved accuracy. We use advanced ML methods to solve the overdetermined system obtained from images obtained at different voltages.
In general, multimodal ML leverages different data types, such as speech and images, to improve task-specific performance [14]. In medical imaging, multimodal data fusion, such as combining positron emission tomography with computed tomography and/or magnetic resonance imaging, enhances lesion quantification [15,16,17,18]. Shared image features from unregistered views enhance classification [19]. A comprehensive summary of multimodal ML developments in medical imaging can be found in [20]. In electron microscopy, combining signals from high-angle annular dark-field and energy-dispersive X-ray spectroscopy facilitates the 3D reconstruction of nanostructures [21], and combining transmission X-ray microscopy with FIB-SEM improves the image quality for shale analysis [22].
In this study, we present a novel ML-based method for a multi-voltage (multiV) FIB tomography dataset of HNPG. To develop this method, we created synthetic multiV images using a Monte Carlo-based method [12]. These simulated multiV data are used to train ML models. We compared different segmentation methods and demonstrated significant segmentation improvements with multiple modalities. In particular, our multimodal ML method outperforms single-modality ML methods in mitigating the shine-through effect.
2 Materials and Methods
2.1 Acquisition of Imaging Data
In this study, we used imaging data obtained from both real HNPG samples and computer-generated synthetic imaging data, closely resembling actual HNPG imaging data, to construct datasets for ML.
2.1.1 Real Samples
An HNPG sample featuring a uniform random network structure with two well-defined ligament sizes of 15 and 110 nm was prepared using the dealloying-coarsening-dealloying method [23]. To improve SEM imaging and distinguish solid and pore phases, epoxy resin infiltration was employed because it provides a dark, homogeneous background during SEM imaging [24]. MultiV FIB tomography of the HNPG sample was performed using a Dual Beam FEI Helios NanoLab G3 system, integrating ASV4 Thermo Fisher Scientific Inc (2017) software [25] for automated tomography control. This software acquires electron- and ion-beam images to monitor milling progress and compensate for image drift resulting from various sources, including charge effects, mechanical stage drift, and thermal-induced drift [26].
To perform drift compensation during FIB tomography, two fiducial markers were prepared and positioned on the side of the region of interest (ROI). Each fiducial marker comprised two perpendicularly intersecting trenches milled on a platinum-deposited area, separate from the ROI. In addition, a ruler system was implemented on top of the ROI to ensure precise slice thickness determination, following a methodology similar to that presented in [27] and optimized for HNPG tomography [3]. This approach involved sputtering a 1-µm carbon layer atop the ROI to smoothen the surface and render the ruler visible (see Fig. 2).
Experimental dataset. A Schematic overview. Dark areas with white crosses represent fiducial structures for drift compensation. B Imaged block face-electron-beam view (BSE, TLD, 2 kV, 50 pA, \(52^\circ\) tilt) and C milling view (ETD, SE, 30 kV, 80 pA, \(0^\circ\) tilt) with (1) ruler structure, (2) e-beam fiducial structure, (3) HNPG infiltrated with epoxy, (4) ion-beam fiducial structure, (5) 30-µm deep trenches, and (6) direction of ion mill (scale bar: 1 µm)
During multiV tomography, each slice was imaged three times with accelerating voltages of 1, 2, and 4 kV at a constant current of 50 pA. This process generated three real FIB tomography image datasets referred to as r-1 kV, r-2 kV, and r-4 kV (where the letter “r” refers to the fact that the data are based on real rather than synthetic, computer-generated microstructures). The SEM image parameters included a resolution of 3072 × 2048 and a horizontal field of view spanning 10 µm, resulting in a pixel size of 3.26 nm. Low-noise imaging was achieved with a dwell time of 30 µs. A through-the-lens detector (TLD) was used to detect back-scattered electrons (BSEs).
In total, 316 slices were milled using an ion beam with an aperture of 80 pA and an accelerating voltage of 30 kV. This dataset yielded a mean slice thickness of \(<d>\,=\,10.16\) nm, with a standard deviation of \(\sigma \,=\,3.12\) nm, aligning closely with the target thickness of 10 nm. Notably, 9% of the slices, characterized by thickness outliers (z-scores of 1.5), were excluded from the thickness calculation analysis.
Owing to instrument constraints, misalignments frequently occur in images captured at different accelerating voltages within and across stacks (see Fig. 3). Consequently, image registration is essential to ensure that the same region is consistently exposed across all voxel groups for images obtained at different accelerating voltages. This registration process comprises two key steps:
Intra-stack image registration
The first step involves aligning the images within each image stack acquired at a specific accelerating voltage (1, 2, and 4 kV). The initial image in each stack serves as a reference. Subsequent images are translated in the (in-plane) x- and y-directions to maximize the correlation between consecutive slices. We used the Fiji plugin [28] ‘Register virtual stack slices’ to perform intra-stack registration.
Inter-stack image registration
After intra-stack registration, images across stacks acquired with different voltages (1, 2, and 4 kV) were aligned considering the image stack acquired at 2-kV accelerating voltage as the reference. Calculating correlations between corresponding slices in 1- and 2-kV stacks enabled translational shifts for optimal alignment. A supplementary fiducial marker, milled on the right side of the sample (Fig. 2), enhanced the inter-stack correlation as no shine-through effect is noticed for the region across different image stacks. Aligning the 4- and 2-kV stacks followed the same procedure. Figure 4 shows an identical slice from each dataset after the registration procedure.
Single raw FIB-SEM slice images of real HNPG at A 1 kV, B 2 kV, and C 4 kV, each featuring a fiducial marker in the xy-plane. Misalignment is evident from the marked rectangles (red), showcasing variations in x- and y-directions between images acquired at different accelerating voltages. Scale bar: 1000 nm
2.1.2 Synthetic Samples
Supervised ML on electron microscopy data, particularly for FIB tomography of HNPG, can be challenging because of the high cost and time involved in acquiring substantial training datasets. Because ground truth images are unavailable for the FIB tomography data of HNPG, we generated synthetic FIB tomography images, as described in [12]. In the first step, virtual initial structures resembling real HNPG structures were generated using the method described in [23]. The leveled-wave model [29] provides a basis for each virtual HNPG hierarchy level. This model starts with a concentration field formed by the superimposition of waves with the same wavelength but with random wave vector directions. The wave vectors have the same magnitude, which dominates the ligament size of the virtual nanoporous network. Subsequently, a level cut was selected for binarization into a pore or solid phase, and virtual nanoporous microstructures were generated after removing the pore phase. Various level cuts can be applied to achieve the desired solid fraction. In the next step, the process of FIB-SEM tomography was simulated on these binary structures by Monte Carlo simulations using the MCXray plugin of Dragonfly software [30]. We generated three datasets using the same virtual initial structure but at different accelerating voltages (1, 2, and 4 kV), similar to the real HNPG data. As shown in Fig. 5, these datasets, named s-1 kV, s-2 kV, and s-4 kV (where “s” refers to “synthetic”), resemble the real HNPG data acquired at different accelerating voltages.
2.2 Machine Learning Architectures for Semantic Segmentation
We used multimodal ML to segment the FIB tomography data semantically. Inspired by [20], we developed three architectures for 3D nanostructure reconstruction, adapted from [12], employing different data fusion techniques:
2.2.1 Early Fusion
In the early fusion architecture (Fig. 6(A)), we fused multimodal images at the input level by channel-wise concatenating single two-dimensional (2D) slices from the s-1 kV, s-2 kV, and s-4 kV datasets. These slices form the input tensor, passing through custom 2D and 3D U-Net models to produce the final segmentation output.
2.2.2 Intermediate Fusion
The intermediate fusion architecture (Fig. 6(B)) contains separate branches for each imaging modality, focusing on images at different accelerating voltages. These images pass through modality-specific subnetworks to extract low-level features, which are then concatenated and classified using a fully connected neural network to yield the final segmentation. This architecture posed challenges because of its size during the optimization process.
2.2.3 Late Fusion
The late fusion architecture (Fig. 6(C)) resembles intermediate fusion, with each branch dedicated to a different modality, initially trained and optimized. These trained ML models estimate the probabilities of each pixel belonging to either the material or pore phase. In the second step, the output probabilities from these optimized branches are collected and concatenated. These concatenated probabilities then train a final neural network layer (ensemble), which, once optimized, produces the ultimate segmentation output.
2.3 ML Model Training Process
We trained all ML architectures on Tesla K80 GPUs. Images were initially cropped into smaller patches (64 \(\times\) 64) with a 32-pixel stride using a sliding window technique. Our networks are trained using a structured approach where data are presented as individual 2D slices, a 3D volumetric stack, or a 2D slice coupled with its neighboring slices for a 2D convolutional neural network (CNN), 2D CNN with adjacent slices, and 3D CNN. We used Dice loss in conjunction with the Adam optimizer, starting with an initial learning rate of 0.0001. If the learning rate did not decrease for ten consecutive training epochs, the learning rate was reduced by a factor of 10. Table 1 shows details of the training parameters, and supplementary (Appendix) Fig. 10 illustrates a typical training and validation data Dice loss curve.
2.4 Data Augmentation
ML requires a large set of training data. When it is difficult to collect sufficient training data, data augmentation can be a powerful method for increasing the data size for ML [31]. Differences in data distributions between synthetic and real datasets and variations stemming from different microscopes emphasize the need for extensive data augmentation in electron microscopy. We applied online data upsampling, where the data size was increased during training by applying different image processing operations such as random flips, image rotations, random croppings, and changes in brightness.
2.5 Evaluation Criteria for 3D Image Reconstruction
2.5.1 Metrics Based on Ground Truth Values
In cases where ground truth data are available as a binary virtual initial structure, e.g., for synthetic datasets, we calculated the following three absolute error metrics:
The first metric, the fraction of misplaced pixels (MP), measures the percentage of pixels whose predicted and ground truth values do not match. This metric was calculated using the following formula:
where TP denotes the number of true positives, TN denotes the number of true negatives, FP denotes the number of false positives, and FN denotes the number of false negatives.
The second metric, the percentage of misplaced gold pixels (MGP), calculates falsely predicted gold pixels using ground truth values. One should be careful while using this metric alone, as FP is not considered in this metric. MGP is calculated as follows:
The third metric, the Dice score (DS), measures the overlapping regions between predicted and ground truth binarized images. It provides a valuable measure when data imbalances exist. DS takes values from 0 to 1, 1 being the best performance indicator. DS is calculated as follows:
We computed the DS for both the material and pore phases and then averaged them to obtain the mean Dice score (MDS).
2.5.2 Metrics Based on Anisotropy in the Absence of Ground Truth Values
When ground truth data are unavailable, e.g., for real HNPG FIB tomography, anisotropy-based metrics are used to assess ML model performance under the assumption of isotropy of the actual nanostructure [12]. These metrics account for the shine-through effect observed in the z-direction. Three metrics are calculated in the x-, y-, and z-directions and compared to evaluate anisotropy based on the three functions. The first metric is based on the two-point correlation function (TPCF), which is the probability of having two points in the same material phase at a given distance from each other in x-, y-, and z-directions. Let this function calculated in the three spatial directions be denoted as \(f^{x}\), \(f^{y}\), and \(f^{z}\). When these functions are uniformly discretized with n data points, we obtain the function values \(f^{x}_i\), \(f^{y}_i\), and \(f^{z}_i\), where i ranges from 1 to n. TPCFs statistically characterize the microstructure in different spatial directions. For isotropic microstructures, identical TPCFs are expected in all spatial directions. Conversely, the differences between the TPCFs in different directions are a measure of the anisotropy of the microstructure. To quantify these differences, we calculated the \(L_2\) differences between the TPCF in x- and z-directions and y- and z-directions. These \(L_2\) differences were averaged to obtain the final anisotropy metric as follows:
A similar anisotropy metric can be defined using the lineal path function (LPF) instead of the TPCF. The LPF measures the probability that two points at a certain distance in a certain direction can be connected by a line fully located in the same phase. The resulting anisotropy metric \(e_{L_2}^\textrm{LPF}\) is calculated analogously to \(e_{L_2}^\textrm{TPCF}\). The third metric, \(e_{L_2}^{D}\), is a similar metric and calculates the average difference of ligament diameters in x- and z-directions and y- and z-directions:
Here, \(D_{ij}\) denotes the calculated average diameter of ligaments in the ij plane with \(i,j \in \{x,y,z\}\). When calculated for a completely isotropic structure, zero errors indicate the best reconstruction performance. However, higher error values for these functions suggest anisotropy in the reconstructed structure. For an underlying actual isotropic microstructure, this indicates a segmentation error.
3 Results and Discussion
3.1 Comparison of Different Multimodal ML Architectures
First, we performed a comparative analysis of the three multimodal ML architectures to identify the most effective segmentation model. We trained all three architectures, as described in Sect. 2.2, using synthetic datasets (s-1 kV, s-2 kV, and s-4 kV). After fine-tuning these models, we evaluated their performance using the metrics outlined in Sect. 2.5.1, leveraging the availability of ground truth data for the synthetic dataset. It is essential to emphasize that the synthetic dataset used for metrics calculation remained entirely isolated from that used during the training process. Our results, presented in Table 2, indicate that the late fusion architecture excelled among the three options.
Notably, the intermediate fusion architecture exhibited significantly poorer metrics than the late fusion architecture. This decline in performance can be attributed to the large size of the ML model, which posed challenges for optimization given the limited training data. MP, MGP, and MDS were significantly less favorable for the more straightforward multimodal architecture, early fusion. In addition, we examined the behavior of the late fusion network on the shine-through effect by substituting the ensemble layer with a voting mechanism. This adaptation revealed weights of 0.5, 0.3, and 0.2 for CNN models trained individually on the s-1 kV, s-2 kV, and s-4 kV datasets, respectively, signifying that the model learns valuable insights from the 2- and 4-kV datasets despite their shine-through effects. On the basis of these comparative findings, we decided to proceed with the late fusion architecture for all subsequent parts of this study.
3.2 Comparison of Segmentation Methods (Synthetic Data)
Subsequently, we performed a comparative analysis, pitting our multimodal ML method (ML-multiV) against the cluster-based k-means clustering algorithm and ML models trained using individual single kV datasets, denoted as ML-singleV. The results are tabulated in Table 3, featuring metrics such as MP, MGP, and MDS for the reconstructed simulated structure. ML-singleV (s-1 kV), ML-singleV (s-2 kV), and ML-singleV (s-4 kV) represent cases where the ML model was trained and tested using only a single kV dataset, s-1 kV, s-2 kV, and s-4 kV, respectively.
Notably, our multimodal ML method, ML-multiV, outperformed all alternative techniques assessed. All ML models surpassed the cluster-based k-means clustering method when applied to their respective individual datasets. The best results were observed for the multimodal ML approach, particularly when considering all three datasets for segmentation. This underscores the distinct advantage gained from the additional information acquired through images captured at various accelerating voltages.
Supplementary (Appendix) Table 7 provides a detailed exploration of anisotropy-based metrics for each segmentation method, reaffirming the trends observed in Table 3. In addition, Fig. 7 offers a visual representation of the segmentation outputs, further supporting the efficacy of our multimodal ML technique.
3.3 Comparison of Performance for Real HNPG Data
We have demonstrated the superior performance of our multimodal ML approach on synthetic datasets created using Monte Carlo simulations of the FIB tomography process. To validate these results on real data, we assessed the segmentation performance of our ML-multiV method on real HNPG FIB tomography data (r-1 kV, r-2 kV, and r-4 kV) using anisotropy-based metrics (see Sect. 2.5.2). The outcomes presented in Table 4 confirm the advantages of our method, ML-multiV, over alternatives when applied to real FIB tomography data. Figure 8 shows a single slice from a segmented real HNPG dataset using different segmentation methods. This figure illustrates the enhanced ability of all ML-based techniques in mitigating the shine-through effect when compared with classical segmentation methods. Note that ML-multiV, on average, exhibits more than 50% improvement in performance based on anisotropy measures compared with k-means clustering. Furthermore, the enhancement over k-means clustering (r-1 kV) in anisotropy measures exceeds 30%.
4 Conclusion
FIB-SEM tomography data are affected by artifacts such as the shine-through effect and the resulting ambiguity in image intensities. These artifacts make it difficult to use cluster-based segmentation methods, such as k-means clustering. More advanced ML-based methods can efficiently suppress the shine-through effect, even when trained only on a single set of synthetic FIB tomography images. The reconstruction performance can be further improved using more than one set of FIB tomography data for the same system. Exploiting this idea, we developed an overdetermined system for FIB-SEM tomography of nanostructured materials such as HNPG. We accomplished this by combining FIB tomography data collected for the same system at different acceleration voltages. The additional data collected in this way can help decide with greater certainty whether certain high-intensity pixels of the SEM image belong to the surface area or are rather shine-through artifacts from deeper layers that should ideally be neglected.
This study demonstrated that a multimodal ML method using imaging data collected with various acceleration voltages can outperform classical segmentation methods using only one dataset (obtained with a single acceleration voltage). We demonstrated this specifically using a multimodal ML architecture that combined imaging information obtained at acceleration voltages of 1, 2, and 4 kV.
Availability of Data and Material
The datasets generated and analyzed for this study can be found in TUHH Open Research repository. DOI: https://doi.org/10.15480/882.8927
References
Liu Y, Zhao T, Ju W, Shi S (2017) Materials discovery and design using machine learning. J Materiomics 3(3):159–177
Fam Y, Sheppard TL, Diaz A, Scherer T, Holler M, Wang W, Wang D, Brenner P, Wittstock A, Grunwaldt J-D (2018) Correlative multiscale 3D imaging of a hierarchical nanoporous gold catalyst by electron, ion and X-ray nanotomography. ChemCatChem 10(13):2858–2867
Shkurmanov A, Krekeler T, Ritter M (2022) Slice thickness optimization for the focused ion beam-scanning electron microscopy 3D tomography of hierarchical nanoporous gold. Nanomanuf Metrol 5(2):112–118
Richert C, Huber N (2018) Skeletonization, geometrical analysis, and finite element modeling of nanoporous gold based on 3D tomography data. Metals 8(4):282
Hu K, Ziehmer M, Wang K, Lilleodden ET (2016) Nanoporous gold: 3D structural analyses of representative volumes and their implications on scaling relations of mechanical behaviour. Phil Mag 96(32–34):3322–3335
Sardhara T, Shkurmanov A, Li Y, Shi S, Cyron CJ, Aydin RC, Ritter M (2023) Role of slice thickness quantification in the 3D reconstruction of FIB tomography data of nanoporous materials. Ultramicroscopy. https://doi.org/10.1016/j.ultramic.2023.113878
Prill T, Schladitz K, Jeulin D, Faessel M, Wieser C (2013) Morphological segmentation of FIB-SEM data of highly porous media. J Microsc 250(2):77–87
Fager C, Röding M, Olsson A, Lorén N, Corswant C, Särkkä A, Olsson E (2020) Optimization of FIB-SEM tomography and reconstruction for soft, porous, and poorly conducting materials. Microsc Microanal 26(4):837–845
Sardhara T, Shkurmanov A, Aydin R, Cyron CJ, Ritter M (2023) Towards an accurate 3D reconstruction of nano-porous structures using fib tomography and monte carlo simulations with machine learning. Microsc Microanal 29(Supplement_1):545–546
Rogge F, Ritter M (2019) Cluster analysis for FIB tomography of nanoporous materials. In: Conference: IMC19, Sydney
Fend C, Moghiseh A, Redenbach C, Schladitz K (2021) Reconstruction of highly porous structures from fib-sem using a deep neural network trained on synthetic images. J Microsc 281(1):16–27
Sardhara T, Aydin RC, Li Y, Piché N, Gauvin R, Cyron CJ, Ritter M (2022) Training deep neural networks to reconstruct nanoporous structures from FIB tomography images using synthetic training data. Front Mater 9:837006
Liu Y, Yang Z, Zou X, Ma S, Liu D, Avdeev M, Shi S (2023) Data quantity governance for machine learning in materials science. Natl Sci Rev 125
Yuhas BP, Goldstein MH, Sejnowski TJ (1989) Integration of acoustic and visual speech signals using neural networks. IEEE Commun Mag 27(11):65–71
Beyer T, Townsend DW, Brun T, Kinahan PE, Charron M, Roddy R, Jerin J, Young J, Byars L, Nutt R (2000) A combined PET/CT scanner for clinical oncology. J Nucl Med 41(8):1369–1379
Bagci U, Udupa JK, Mendhiratta N, Foster B, Xu Z, Yao J, Chen X, Mollura DJ (2013) Joint segmentation of anatomical and functional images: Applications in quantification of lesions from PET, PET-CT, MRI-PET, and MRI-PET-CT images. Med Image Anal 17(8):929–945
Lian C, Ruan S, Denœux T, Li H, Vera P (2018) Joint tumor segmentation in PET-CT images using co-clustering and fusion based on belief functions. IEEE Trans Image Process 28(2):755–766
Suk H-I, Lee S-W, Shen D, Initiative ADN et al (2014) Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage 101:569–582
Carneiro G, Nascimento J, Bradley AP (2015) Unregistered multiview mammogram analysis with pre-trained deep learning models. In: conference on medical image computing and computer-assisted intervention, pp 652–660. Springer
Guo Z, Li X, Huang H, Guo N, Li Q (2019) Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans Radiat Plasma Med Sci 3(2):162–169
Huber R, Haberfehlner G, Holler M, Kothleitner G, Bredies K (2019) Total generalized variation regularization for multi-modal electron tomography. Nanoscale 11(12):5617–5632
Anderson TI, Vega B, Kovscek AR (2020) Multimodal imaging and machine learning to enhance microscope images of shale. Comput Geosci 145:104593
Shi S, Li Y, Ngo-Dinh B-N, Markmann J, Weissmüller J (2021) Scaling behavior of stiffness and strength of hierarchical network nanomaterials. Science 371(6533):1026–1033
Peña B, Owen GR, Dettelbach K, Berlinguette C (2018) Spin-coated epoxy resin embedding technique enables facile SEM/FIB thickness determination of porous metal oxide ultra-thin films. J Microsc 270(3):302–308
Thermo Fisher Scientific Inc: Auto slice and view 4.0 [computer software] (2017). Version: 4.1.0.1196. Accessed 2022-07-13
Lepinay K, Lorut F (2013) Three-dimensional semiconductor device investigation using focused ion beam and scanning electron microscopy imaging (FIB/SEM tomography). Microsc Microanal 19(1):85–92
Jones H, Mingard K, Cox D (2014) Investigation of slice thickness and shape milled by a focused ion beam for three-dimensional reconstruction of microstructures. Ultramicroscopy 139:20–28
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682
Soyarslan C, Bargmann S, Pradas M, Weissmüller J (2018) 3D stochastic bicontinuous microstructures: generation, topology and elasticity. Acta Mater 149:326–340
Object Research Systems (ORS) Inc, C. Montreal: Dragonfly 3.6 [computer software] (2018)
Perez L, Wang J (2017) The effectiveness of data augmentation in image classification using deep learning. arXiv preprint arXiv:1712.04621
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, October 5–9, 2015, Proceedings, Part III 18, pp 234–241. Springer
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Medical image computing and computer-assisted intervention–MICCAI 2016: 19th international conference, Athens, Greece, October 17–21, 2016, Proceedings, Part II 19, pp. 424–432. Springer
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - SFB 986 - Project number 192346071.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. Sardhara, Aydin, Cyron, and Ritter contributed to the conception and design of the study. Shkurmanov acquired FIB-SEM images of HNPG and calculated slice thicknesses. Li generated the LWM database. Shi and Riedel prepared the HNPG sample. Sardhara wrote the first draft of the manuscript. All authors contributed to the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix 1: Machine Learning Architecture
We designed custom U-Net-based architectures for multimodal ML models (Fig. 6). These architectures were tailored for each fusion method and differed in their approach to processing image data. In all U-Net architectures [32, 33], we incorporated customizations such as residual connections, padding, and the number of encoding/decoding blocks (Fig. 9). After evaluating the performance of various architectures for different fusion techniques, we selected the following:
In the early fusion approach, we opted for a 3D U-Net model comprising four encoding blocks with residual connections.
In the case of intermediate fusion, we narrowed our focus to a 2D CNN and a 2D CNN with adjacent slices because of the significant size of the multimodal ML model. To our surprise, the 2D CNN proved to be the most effective, probably because of the constraints of the limited dataset size and the overall large model size after intermediate fusion. Consequently, we adopted a 2D U-Net model that features two encoding blocks with residual connections. The fully connected layer block incorporated two convolutional layers: The first expanded output channels to 64, followed by a second output classifier layer with a kernel size of 1.
For the late fusion approach, we trained and compared the 2D CNN, 2D CNN with adjacent slices, and 3D CNN architectures, with the 2D CNN featuring adjacent slices outperforming the others. This approach employed seven adjacent slices, four encoding blocks, and residual connections and included an ensemble block with a convolutional layer and a kernel size of 1.
Appendix 2: Training Curves
Figure 10 depicts training and validation Dice loss curves. These curves display the mean Dice loss over epochs, with the blue line representing training scores and the orange line representing validation scores. We used data augmentation during training, which made the training dataset more complex. Consequently, validation Dice loss values were consistently lower than training Dice loss values in all curves.
Appendix 3: Effect of 3D Reconstruction on Material Properties (Solid Fraction)
The solid fraction (\(\phi\)), which is a critical property in materials science, quantifies the volume of the solid phase within a structure. It significantly influences mechanical and optical properties, underscoring the importance of its accurate measurement, which relies on precise 3D reconstruction. The solid fraction can be calculated from the binary structure as follows:
where \(N_m\) represents voxels within phase m and \(N_\textrm{total}\) denotes the total voxel count. Synthetic data with ground truth allow us to observe the impact of reconstruction on the solid fraction. Table 5 shows that the ML-based methods yield similar solid fractions, while with k-means clustering, the solid fraction increases with increasing voltage, indicating its struggle with the shine-through effect.
Real FIB-SEM image data (r-1 kV, r-2 kV, and r-4 kV) lack ground truth, limiting direct comparison. Nevertheless, Table 6 shows that ML-singleV (r-1 kV), ML-singleV (r-2 kV), and ML-multiV exhibit solid fractions indicative of realistic hierarchical nanoporous materials.
Appendix 4: Additional Results Using Anisotropy-Based Error Measures for Synthetic Data
In this section, we computed anisotropy-based errors as detailed in Sect. 2.5.2 to compare the performance of different segmentation methods. Table 7 presents the comprehensive metrics for the synthetic datasets including for the original dataset, serving as reference values for evaluating the outcomes obtained from different methods.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sardhara, T., Shkurmanov, A., Li, Y. et al. Enhancing 3D Reconstruction Accuracy of FIB Tomography Data Using Multi-voltage Images and Multimodal Machine Learning. Nanomanuf Metrol 7, 4 (2024). https://doi.org/10.1007/s41871-024-00223-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41871-024-00223-y