Abstract
Cyclaneusma needle cast (CNC) is a needle disease which caused deterioration of vitality and reduction in the growth of pines. The disease is caused by the ascomycetous fungus Cyclaneusma minus, which has two well-described morphotypes; C. minus simile and C. minus verum. The distribution and host range of C. minus simile and verum was determined from needle samples and isolates collected throughout Slovakia from 2014 to 2020. Samples from 111 localities, 11 pine host species and 245 trees collected in different types of planting were analysed. It was found, that both morphotypes are present, but C. minus verum is predominantly responsible for CNC in urban and forest plantings in Slovakia. C. minus verum was positively detected in more than 88% of collected samples, whereas C. minus simile was only in four samples from three localities. Morphotype-specific primers were sufficiently sensitive even for new pine-host species. The host range of C. minus simile and C. minus verum was enriched worldwide. C. minus verum was observed in nine host species, whereas C. minus simile was identified only in one. Cyclaneusma niveum was also recorded and its presence was confirmed through DNA sequencing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyclaneusma needle cast (CNC) is a foliar disease of Pinus species which infects needles of Austrian (P. nigra Arn.), Eastern white (P. strobus L.), Monterey (P. radiata D. Don), ponderosa pine (P. ponderosa Douglas ex C. Lawson), Scots (P. sylvestris L.) and mountain pine (P. mugo Turra). It is particularly common and detrimental to the growth of P. radiata plantation forests in New Zealand and Australia (Watt et al. 2012a, b), but is also found in Asia (Ying-Ren 2012), Europe (Sieber et al. 1999; Markovskaja et al. 2016; Kowalski 1988; Dubach et al. 2022), USA (Merrill and Wenner 1996) and South Africa (Crous et al. 1990).
Two Cyclaneusma species have been formally described, Cyclaneusma minus (Butin) Di Cosmo, Peredo and Minter and Cyclaneusma niveum (Pers.) Di Cosmo, Peredo and Minter (Di Cosmo et al. 1983). Both Cyclaneusma species are found in Slovakia (Ivanová 2015; Pastirčáková et al. 2014; Hečková 2016; Zúbrik et al. 2019). Cyclaneusma niveum was found on the old needles of P. nigra in urban greenery (Ivanová 2015; Pastirčáková et al. 2014), and also recorded on P. sylvestris needles (Pastirčáková et al. 2014). Cyclaneusma minus has been recorded in pine forests in Slovakia, but with sporadic occurrence (Zúbrik et al. 2019).
Cyclaneusma minus was one of the most abundant pathogens in Scots pine needles, detected by Illumina sequencing, a culture‐independent method used to examine fungal communities. It was frequent in samples showing CNC symptoms, but also very frequent in symptomless needles (Behnke-Borowczyk et al. 2019). Cyclaneusma minus can behave as an endophyte and as pathogen on the same host species depending on the host genotype (Sieber et al. 1999; Bulman 1993; McDougal et al. 2012). These results suggest that this fungal species is a member of the “mutualism–parasitism continuum” where different host and fungal genotypes and broad functional host groupings result in different host responses within the host–fungus associations affected by abiotic conditions or by the presence of other pathogens (Mandyam and Jumpponen 2015). The first report documenting morphological differences in C. minus cultures was recorded by Karadzic (1981). Subsequently, this variation was also observed in New Zealand (Bulman and Gadgil 2001, Dick et al. 2001). Morphological differences in mycelium colour, colony texture, pigmentation, sporulation and growth in vitro, and measurements of apothecial length have been used to separate C. minus into two different morphotypes. These morphotypes have been named C. minus simile (Cm simile) and C. minus verum (Cm verum) (Bulman and Gadgil 2001; Dick et al. 2001). Results of phylogenetic analysis with multiple gene regions have shown that the two morphotypes are likely to be distinct species which differ by an indel of 192 bp in the 18S rDNA (Prihatini et al. 2014). In the multigene phylogenetic analysis conducted by Prihatini et al. (2014) on two C. minus morphotypes, isolates from New Zealand and Australia were examined, along with three additional isolates from other continents: two from Europe (Germany) and one from Africa (Kenya). The isolates from New Zealand and Australia included both morphotypes, while the three isolates from outside Australasia were all identified as Cm verum.
Hunter et al. (2016) developed a molecular method for the rapid diagnosis and characterization of these C. minus morphotypes, using specific primers for both conventional and real-time PCR, applicable to cultures and directly to infected needles. This method was tested on DNA from fungal species isolated from P. radiata as well as P. radiata DNA (from needles) collected in New Zealand plantation forests. In that study, a third morphotype was recognized, but could not be characterized using these molecular tools. Recent research has identified the presence of a third C. minus morphotype in New Zealand, where genome sequencing and effector analysis, in addition to traditional mycological techniques, demonstrated clear differences among the three morphotypes, suggesting the need for formal species descriptions for each of the morphotypes (Tarallo et al. 2023). In this study, the aim was to: (i) evaluate the efficiency and sensitivity of specific primers in detecting the Cm simile and Cm verum morphotypes in different pine species; (ii) identify the morphotype responsible for CNC in Slovakia; and (iii) establish the distribution and host range of both morphotypes across Slovakia.
Material and methods
Sample collection and disease incidence
Symptomatic pine needles were taken from 2014 to 2020 across Slovakia to cover the entire territory of the country. The needles were collected from hanging branches or from the ground beneath trees and were examined for typical symptoms of CNC. One sample consisted of a pooled collection of at least 30 symptomatic needles, each with fruiting bodies, from a single pine tree. Each sample was sealed and labelled in a separate bag and stored in a freezer at -20 °C until processing.
Sampling sites
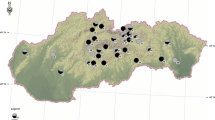
Samples from 111 localities collected in different types of planting taken randomly across Slovakia were analysed. In total 245 trees were sampled which included 201 needle samples from urban greenery, 21 from arboreta, 18 from forest plantation and five samples came from natural forest regeneration (Table S1). Symptoms present in trees, with positive identifications for Cyclaneusma spp., included yellow-green mottling or marbling of the pine needles, eventually the presence of necrotic needles on pine branches, with typical white fungal fruiting bodies, nicely recognizable in wet weather. The fruiting bodies (apothecia) were roughly rectangular or elliptical, waxy, reddish-brown when young and later become the same colour as the needle surface. Mature apothecia swell when moist and the half-lids formed by the hinged epidermis are pushed back, exposing the slightly convex, straw-coloured, spore-bearing layer (Bulman and Gadgil 2001). The fruiting bodies were predominantly on the base of the needle, but if disease was severe, also along the needle.
Fungal isolation
The needles with typical fruiting bodies (roughly rectangular to elliptical, waxy, reddish-brown later become the same colour as the needle surface (Bulman and Gadgil 2001) were surface-sterilized according to Sieber (1989). Needles were immersed in 95% ethanol for 1 min., 4% NaOCl for 5 min., 95% ethanol for 30 s., rinsed in sterile H2O and dried on sterile filter paper for a few minutes. They were transferred to Petri dish with filter paper (wetted with sterile distilled water) for 24 h. Subsequently, fruiting bodies with a small portion of needle approximately 0.5 mm were excised under a stereo microscope and transferred to Petri dishes with malt extract agar (MEA, Roth GmbH, Germany 33.6 g/l) medium supplemented with antibiotic (streptomycin sulphate, 100 mg/l). Petri dishes were incubated in a cultivation chamber at 20 °C in the dark for 10–14 days. Isolates obtained in this study were first used for DNA extractions and sequencing. After these DNA sequence-based identifications were confirmed, the isolates were stored in the culture collection at the Institute of Forest Ecology of the Slovak Academy of Sciences (IFE SAS), within the Department of Plant Pathology and Mycology in Nitra, Slovakia. Additionally, herbarium specimens were deposited in the Plant Pathology Herbarium (NR) at IFE SAS once sample processing was finalized.
DNA extraction
DNA was extracted from fungal fruiting bodies or mycelial cultures. One sample comprised at least 10–15 fruiting bodies, each accompanied by a small tissue section from needles about 3–5 mm in length. These were then transferred to a microtube and stored at -20 °C in a freezer for DNA extraction. Cultures isolated from symptomatic needles were also used for DNA analysis. Extraction of DNA from mycelia of pure fungal cultures was performed after culturing for 2 weeks on 3% MEA. The tissues (100 mg) were ground using a sterile mortar and pestle and liquid nitrogen. The DNA extractions were performed using a E.Z.N.A. Fungal DNA Mini kit (Omega Bio-Tek Inc., Norcross, GA USA) according to the instructions provided by the manufacturer. DNA was dissolved in 60 µl elution buffer and stored in a freezer at -20 °C.
Species detection using morphotype-specific PCR
The presence of C. minus in extracted DNA from fungal fruiting bodies on needles and cultures was detected by conventional PCR using morphotype-specific primers.
C. minus verum morphotype was detected using primers Cm verum specific ITS: Cm verum F (CCGGGCCTTCGGGCCTAC)/Cm verum R (CAGGCACCAACCCAGGC) and morphotype C. minus simile was detected using primers: Cm simile F (CCGGGCCTTATGGTCCGC)/Cm simile R (CAGGCGCCAGCCCAGCG) (Hunter et al. 2016). Each DNA sample was analysed with both pairs of primers. The PCR reaction was prepared in a total volume 20 µl containing of 2 ng / μl DNA template, forward and reverse primer (10 pmol / μl), 5 × HOT FIREPol®BlendMaster Mix (SolisBioDyne, Tartu, Estonia), sterile molecular grade deionized water. A DNA sample of Cm verum and Cm simile identified in our previous analyses was used as a positive control, while the negative control contained PCR mastermix with sterile deionized water. An initial denaturation step at 95 °C for 15 min., was followed by 35 amplification cycles of denaturation at 95 °C for 15 s, annealing at 63 °C for 15 s and extension at 72 °C for 15 s. The thermal cycling finished by a final extension at 72° for 10 min (Hunter et al. 2016). PCR products were separated by 80 V electrophoresis for approximately 1 h in a 0.8% (w / v) agarose gel using Simply Safe dye (EURx, Poznan, Poland) and the gel was visualized under UV light. We used a 100 bp marker (Solis BioDyne, Tartu, Estonia) to determine the amplicon size. The thermal cycling was carried out in a T-professional thermocycler (Biometra, Göttingen, Germany).
DNA sequencing of cultures
Pure cultures of Cyclaneusma species grown from cultivation of fruiting bodies on surface sterilized needles were analysed by DNA sequencing. For the identification of fungal cultures, we amplified the ITS region using the universal primers ITS1F and ITS4 according to White et al. (1990) and Gardes and Bruns (1993). The PCR reaction was performed as described in the species detection section. PCR conditions included an initial denaturation at 95 °C for 14 min, followed by 13 cycles of denaturation at 95 °C for 35 s, annealing at 55 °C for 55 s, and elongation at 72 °C for 45 s; followed by a further 13 cycles of denaturation at 95 °C for 35 s, annealing at 55 °C for 55 s, and elongation at 72 °C for 2 min, and then lastly 9 cycles with the same condition for denaturation and annealing, with a longer elongation of 3 min. A final extension was carried out at 72 °C for 10 min. Prior to sequencing, target fragments were directly purified using a PCR Purification Kit (Qiagen GmbH, Hilden, Germany). Each amplified product was diluted in 30 µl of sterile distilled water. Sequencing was performed at SEQme Ltd. (Dobříš, Czech Republic) using a ABI3130xl sequencer (Applied Biosystems). The retrieved sequences were compared by BLAST (Basic Local Alignment Search Tool, available at http:// www.ncbi.nlm.nih.gov/genbank/). Sequences were deposited in NCBI GenBank. GenBank accession numbers are available in Tables 2 and S1.
Results
Cyclaneusma minus morphotypes, host range and incidence
DNA was extracted from 245 needle samples of eleven Pinus species, and tested for the presence of C. minus verum or simile using the molecular tools described in Hunter et al. (2016). Morphotype C. minus verum was recorded in 201 needle samples from 98 localities (> 82%) (Table 1). Both C. minus verum and simile produced the expected 390 bp amplicon on an agarose gel (Hunter et al. 2016).
We recorded the highest percentage detection for the C. minus verum in samples from natural regeneration, where all 5 samples (100%) yielded positive results for the detection of this morphotype (Table S1). In urban conditions, the proportion of positive results for C. minus verum detection was high at 83% (167 samples). Similarly, for other types of planting, this percentage was high, 77% (14 from 18 samples) and 71% (15 from 21 samples) for forest plantation and arboretum samples, respectively (Table S1).
Detection of the Cyclaneusma minus verum showed that its morphotype had a wide host range on Pinus species across Slovakian sites (Table 1). The presence of this morphotype was noted in nine of the eleven hosts. The most common host detections were on P. sylvestris (85%), P. nigra (83%) and P. mugo (83%). One or two positive samples were found in P. coulteri, P. densiflora, P. jeffreyi, P. ponderosa, P. strobus and P. uncinata. Only P. cembra and P. contorta tested negative for Cyclaneusma presence.
Cyclaneusma minus simile was only detected in 4 samples from three localities, two of which (Nitra, Mochovce) are located in the west region and one (Litava) in the middle part of the country. In 2 cases, these samples were collected from a forest plantation and 2 from an urban environment. All C. minus simile positive samples were from the same host P. nigra. Despite the fact that the detection of C. minus simile was subjected to all 245 samples, this morphotype was not detected on any other hosts (Table 1). However, the incidence of this morphotype was only in 1.6% of all needle samples; a much lower detection rate compared with C. minus verum. All four C. minus simile samples were recorded with positive detection of both the simile and verum morphotypes.
Assessment of Cyclaneusma diversity using a culture-based approach
Out of the seven isolated cultures, five tested positive for C. minus verum using specific primers for Cyclaneusma morphotypes, while none were identified as C. minus simile. Two isolates yielded negative results for both Cyclaneusma morphotype-specific primers. DNA sequencing and Blast analysis of ITS sequences from DNA of these cultures revealed four different fungal species: C. niveum (n = 3), C. minus (n = 2), Biscogniauxia nummularia (n = 1) and Microsphaeropsis olivacea (n = 1) (Table 2). Cultures SY 274 and SY 279 were tested positive for C. minus verum using the morphotype primers, but DNA sequencing showed that these were not Cyclaneusma spp. at all and were instead identified as Biscogniauxia nummularia and Microsphaeropsis olivacea. Sequence analysis confirmed the identity of C. niveum from three cultures (SY 271, SY 272 and SY 278), despite all cultures testing positive using Cyclaneusma morphotype specific primers for C. minus verum.
Two cultures, SY 14 and SY 15, gave no signal using Cyclaneusma morphotype specific primers either for C. minus verum or for C. minus simile, however DNA sequencing and BLAST analysis showed that these isolates were C. minus verum, which gave 100% similarity to the Cyclaneusma minus type strain CBS 496.76 (Genbank accession number NR153910) published by Hunter et al. (2016).
All C. niveum cultures were isolated from P. nigra and the two cultures identified as C. minus verum were isolated from P. sylvestris.
Discussion
In this study, C. minus morphotype-specific primers described by Hunter et al. (2016) were tested using a large number of samples from conifer needles (245 needle samples and 7 cultures) collected from 11 pine species. This study is the first to provide results about the incidence of the known C. minus verum and simile morphotypes in Europe, specifically their geographical distribution and host range in Slovakia.
We confirmed the presence of both Cyclaneusma morphotypes using conventional PCR, which to date has been reported only based on morphological measurements from Slovakia. Even for C. minus only simple data regarding the presence were published without any detailed information about the distribution or host range (Hečková 2016; Zúbrik et al. 2019).
In this work, using these molecular tools (Hunter et al. 2016) for the detection of C. minus morphotypes, 44 needle samples from a total of 245 analysed (18%) gave negative results for both C. minus morphotypes. The primers (Cm simile and Cm verum ITS specific) have high sensitivity; the limit of detection using conventional PCR was 1 and 10 pg, respectively. Cyclaneusma minus can be detected from pine needles by growing out the fungus, and this approach has been used successfully in both symptomatic and asymptomatic needles (Hunter et al. 2016). The negative results obtained in this study may indicate that the extracted DNA could have a low titre of Cyclaneusma species within the needle. Hunter et al. (2016) often used two rounds of PCR to detect C. minus in infected needles. That study also indicated that the DNA extraction method may not be efficient enough for detection of this pathogen from needle tissue or co-extracted contaminants might affect the PCR. In this study, we did not need to use two rounds of PCR to detect from pine needles and showed higher rates of detection from pine needles (84%) success rate, compared to 50% success rate by Hunter et al 2016). These fungi are known to exist as asymptomatic pathogens and potentially as endophytes, with the fungal-to-plant DNA ratio remaining unknown (Helander et al. 1994; Kowalski 1993; Sieber et al. 1999). Alternatively, variations in the ITS region could occur in both C. minus simile and C. minus verum. This has already been observed in one isolate from the New Zealand collection, which showed DNA sequence variation in the ITS region for both morphotypes. The C. minus simile and C. minus verum specific primers were not effective with this isolate (Hunter et al. 2016), and a later study described this isolate as an alternative morphotype named ‘novus’ (Tarallo et al. 2023). Further sequencing of DNA regions could reveal more variability and diversification within C. minus populations in Europe.
A high level of divergence in detection was observed between the two C. minus morphotypes. Cyclaneusma minus verum was identified in more than 82% of collected samples, despite the C. minus simile specific PCR assay reportedly being ten-fold more sensitive than the C. minus verum assay (Hunter et al. 2016). In this study C. minus simile was detected only in four needle samples (1.6% of all samples) from three localities using these primers. Furthermore, from the five cultured Cyclaneusma isolates, none were identified as C. minus simile. Thus C. minus verum appears to be more widespread in Slovakia. Prihatini et al. (2014) included three isolates of C. minus from locations other than New Zealand and Australia (and a total of 37 Cyclaneusma species) in the phylogenetic study of Cyclaneusma species. These isolates (two isolates from Germany and one from Kenya) without host specification were identified as C. minus verum. Other samples analysed from Czech Republic and Poland were identified as C. minus verum as well (data not shown). Furthermore, in another study from Europe using these morphotype specific primers with several pine samples only C. minus verum was identified, as highlighted in Hunter et al. (2016). These existing results describing C. minus morphotype distribution suggest that C. minus verum is more widespread and dominant in Europe, however, more detailed surveys throughout Europe are needed. The opposite pattern of distribution was observed in New Zealand, in the Southern hemisphere, where recently within a large dataset of samples (120 isolates and 18 needle samples) C. minus verum was less common (detected in 11% of needle samples and 21% of cultures by Hunter et al. 2016). However, an earlier study conducted from 1977 to 1983 identified C. minus verum as the most prevalent morphotype in New Zealand (Bulman and Gadgil 2001; Dick et al 2001). This suggests that the dominance of morphotypes in New Zealand may have shifted over the years, or advancements in detection methods have enhanced our ability to identify these pathogens. The possibility of a similar scenario, where a predominance of morphotypes occurs in Slovakia, or more broadly in Europe, remains uncertain. Various factors could influence pathogen behavior, including host genotype, environmental conditions, and endophytes, as well as elements related to human activity, such as survey methodology and sampling practices (Prihatini et al. 2014).
Detection of C. minus in various hosts indicate that C. minus colonizes a diverse range of Pinus species (Butin 1973; Millar and Minter 1980; Lundquist 1986, 1987; Helander et al. 1994; Merrill and Wenner 1996; Sieber et al. 1999; Ganley et al. 2015; Dubach et al. 2022). Based on phylogenetic analysis, the two C. minus morphotypes, verum and simile, belong to two different species (Prihatini et al. 2014; Tarallo et al. 2023). In the present study, we have expanded and specified the host range for the morphotypes C. minus verum and C. minus simile and contributed to the increasing knowledge about the relationship between host species and Cyclaneusma morphotypes. Both C. minus morphotypes were recorded on P. radiata in New Zealand (Dick et al. 2001; Prihatini et al. 2015; Hunter et al. 2016). Similarly, both C. minus verum and C. minus simile were identified on P. nigra in this study, which was the most inspected host species in our collection. This host was introduced in Slovakia, and extensively planted in both urban and forest environments during the 1980s and 1990s (Turis and Valachovič 2014). The widespread cultivation of this species in areas not ideally suited to its requirements may increase its susceptibility to diseases in Slovakia. This was confirmed by other studies where P. nigra was found to be an important host of pathogens such as Lophodermium seditiosum, L. pinastri, Dothistroma septosporum and D. pini (Jánošíková-Hečková et al. 2018; Ondrušková et al. 2023). The susceptibility of P. nigra to both Cyclaneusma species was previously recorded also in New Zealand (Butin 1973).
The morphotype C. minus simile was reported in New Zealand on P. radiata and P. strobus (Bulman and Gadgil 2001; Hunter et al. 2016), and although Cyclaneusma species have been detected on other pine hosts in New Zealand, it is not certain yet which morphotypes these strains belong to (R. McDougal, Scion, unpublished data). In this study, we have only detected C. minus simile on P. nigra in Slovakia. These findings triggered speculation regarding the potential host specificity of C. minus simile in Slovakia. However, further research is required to confirm this, which would involve expanding the sample size across various host species and environmental conditions.
Morphotype C. minus verum was recorded on nine hosts in this study, expanding the known host range of C. minus by two to include P. coulteri and P. densiflora. Based on the wider host range and higher incidence, we can speculate that C. minus verum is more easily adaptable to different conditions of the environment.
By DNA sequencing, C. niveum was identified, and three cultures were isolated from P. nigra needles. Cyclaneusma niveum has been previously identified in this host tree species from urban greenery microscopically and isolated in culture (Ivanová 2015; Pastirčáková et al. 2014), but without molecular confirmation. Although Cyclaneusma species are distinguishable based on microscopic morphological signs, such as the size of ascomata, asci and ascospores, only in ascomatal and ascospore length are the two species well differentiated (Minter and Dudka 1996; Bulman and Gadgil 2001; Dick et al. 2001). Multigene phylogenetic analyses of Cyclaneusma species of five gene regions showed not only that two morphotypes of C. minus belong to different species, but further that C. minus verum was more closely related to C. niveum than to C. minus simile (Prihatini et al. 2014).
Conventional PCR with C. minus morphotype-specific primers was previously determined to be most specific at an annealing temperature of 63 °C and cycle threshold of 35 cycles as only the C. minus simile and verum positive controls amplified at these conditions (Hunter et al. 2016). In that study, PCR specificity was tested using primers against DNA from a range of fungi and oomycetes associated with P. radiata, as well as P. radiata DNA itself (Hunter et al. 2016). However, some cross-amplification did occur at an annealing temperature of 60 °C (lower than the optimal 63 °C) or late in PCR cycling (i.e., cycle threshold values greater than 35) indicating the need to adhere to stringent cycling conditions to maintain specificity. Further, adding both C. minus verum and simile DNA as positive controls for each assay is also useful (Hunter et al. 2016). During our work, all the described PCR conditions were maintained for each completed PCR, expecting the high level of repeatability for the described results. Testing the specificity of these primers by Hunter et al. (2016), as well as an isolate of C. niveum (the type strain, CBS 496.73) was also included with negative detection. The three C. niveum cultures, obtained in this study, gave positive results with morphotype specific primers for C. minus verum, despite these primers testing negative with C. niveum type-strain DNA by Hunter et al. (2016). The morphotype specific primers were designed for the ITS DNA region based on variations observed between the two morphotypes (Hunter et al. 2016), and this DNA region was used for the identification of Cyclaneusma cultures by sequencing. There was a single nucleotide difference in primer sequences of Cm verum F compared to our C. niveum cultures. The sequence of Cm verum R was identical in all ITS sequence of our C. niveum cultures. It is interesting to note that C. niveum is identified from P. radiata at a much lower frequency than C. minus in New Zealand, and New Zealand isolates were not available for testing (R. McDougal, Scion, unpublished data) and this is appearing to be in contrast to the situation in Slovakia where genetic diversity is likely greater.
In this study we also observed additional cross-reactivity of these primers with fungi other than Cyclaneusma species. Two isolates that gave positive results with Cm verum primer set, were identified as Biscogniauxia nummularia and Microsphaeropsis olivacea using ITS PCR and DNA sequencing. These fungi were not used in the specificity testing by Hunter et al. (2016). This highlights the importance of doing specific checks with diagnostics tools that have been published to ensure that they are fit for use with the samples and typical fungi populations that might be encountered. This is not always easy to anticipate however and DNA sequencing to check identifications is a good back up to using species-specific primers.
To understand the impact of this genus of plant pathogens on pine forests it will be necessary to investigate the distribution and genetic diversity in more detail for these Cyclaneusma species, including C. niveum, in Slovakia and Europe more broadly.
Conclusion
The results of the present study contribute new Cyclaneusma needle cast research describing the distribution and host range of two Cyclaneusma minus morphotypes from new regions in Slovakia. This expands on current knowledge about diversity of Cyclaneusma species in Europe. Moreover, further testing of the morphotype-specific primers revealed further variability in Cyclaneusma species. We found that contrasting results were found between New Zealand and Slovakia when determining the prevalence of C. minus verum compared to simile. It would be beneficial to examine severity of disease caused by these morphotypes to understand the impact on Pinus spp. In addition, testing of Slovak isolates of C. niveum points to the possibility of diversified populations of C. niveum in Europe. Together with the recent description of a third morphotype of Cyclaneusma in New Zealand and the use of genomics to examine these, suggests further diversity among Cyclaneusma populations that certainly warrants further detailed genetic studies either at the multigene or genomic level.
For rapid diagnosis of C. minus morphotypes, we suggest combined amplification of the rDNA ITS with DNA sequencing. This study also highlights the need to work collaboratively with other forest pathology researchers to design and develop the new diagnostics tools. This, together with a comprehensive genetic study comprising a larger sample size and geographical scale of Cyclaneusma isolates is recommended, to help improve our understanding of CNC and its impacts on conifer trees.
References
Behnke-Borowczyk J, Kwaśna H, Kulawinek B (2019) Fungi associated with Cyclaneusma needle cast in Scots pine in the west of Poland. For Path 49:e12487. https://doi.org/10.1111/efp.12487
Bulman LS (1993) Cyclaneusma needle-cast and Dothistroma needle blight in NZ pine plantations. N Z For 38(2):21–24
Bulman LS, Gadgil PD (2001) Cyclaneusma needle cast in New Zealand. For Res Bull 222:12–19
Butin H (1973) Morphologische und taxonomische Untersuchungen an Naemacyclus niveus (Pers. ex Fr.) Fuck. ex Sacc. und verwandten Arten. Eur J For Path 3:146–163. https://doi.org/10.1111/j.1439-0329.1973.tb00389.x
Crous PW, Wingfield MJ, Swart WJ (1990) Shoot and needle diseases of Pinus spp. in South Africa. South Afr For J 154:60–66. https://doi.org/10.1080/00382167.1990.9629054
Dick MA, Somerville JG, Gadgil PD (2001) Variability in the fungal population. In: Bulman LS and Gadgil PD (eds.), Cyclaneusma needle-cast in New Zealand, vol 222. Forest Research Bulletin, pp 12–19
DiCosmo F, Peredo H, Minter DW (1983) Cyclaneusma gen. nov., Naemacyclus and Lasiostictis, a nomenclatural problem resolved. Eur J For Path 13:206–212. https://doi.org/10.1111/j.1439-0329.1983.tb00119.x
Dubach V, Queloz V, Stroheker S (2022) Needle and shoot diseases of pine. Swiss Federal Inst WSL Fact Sheet 70:12
Ganley RJ, Hargreaves C, Donaldson LA (2015) Detection of asymptomatic fungal microorganisms in Pinus radiata tissue culture material. N Z J For Sci 45:11. https://doi.org/10.1186/s40490-015-0042-y
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Hečková Z (2016) Damage to Pinus sp. caused by microscopic fungi in selected areas of Slovakia. Mykol Listy Praha 135:77–78
Helander ML, Sieber TN, Petrini O, Neuvonen S (1994) Endophytic fungi in Scots pine needle spatial variation and consequences of simulated acid rain. Can J Botany 72:1108–1113
Hunter S, Glen M, McDougal R (2016) Molecular tools for differentiating Cyclaneusma minus morphotypes and assessing their distribution in Pinus radiata forests in New Zealand. N Z J For Sci 46:2. https://doi.org/10.1186/s40490-016-0080-0
Ivanová H (2015) Fungi associated with a decline of Pinus nigra in urban greenery. Acta Fytotech Zootech 18(2):36–43. https://doi.org/10.13140/RG.2.1.1496.5606
Jánošíková-Hečková Z, Ondrušková E, Barta M, Ostrovský R, Kádasi Horáková M, Pastirčáková K, Kobza M, Adamčíková K (2018) The hosts and geographic range of Dothistroma needle blight in Slovakia. For Pathol 48:e12421. https://doi.org/10.1111/efp.12421
Karadzic D (1981) Infection of Pinus sylvestris by Naemacyclus minor. In: Millar CS (ed) Current Research on Conifer Needle Diseases. Proceedings of the international union of forestry research organisations working party on needle diseases conference. Aberdeen University, Aberdeen, pp 90–101
Kowalski T (1988) Cyclaneusma (Naemacyclus) minus on Pinus sylvestris. Eur J For Path 18(3–4):176–183. https://doi.org/10.1111/j.1439-0329.1988.tb00916.x
Kowalski T (1993) Fungi in living symptomless needles of Pinus sylvestris with respect to some observed disease processes. J Phytopathol 139:129–145
Lundquist JE (1986) Fungi associated with Pinus in South Africa Part I. The Transvaal. South Afr for J 138(1):1–14. https://doi.org/10.1080/00382167.1986.9630036
Lundquist JE (1987) Fungi associated with Pinus in South Africa. Part II. The Cape. South Afr For J 140(1):4–15. https://doi.org/10.1080/00382167.1987.9630063
Mandyam KG, Jumpponen A (2015) Mutualism–parasitism paradigm synthesized from results of root-endophyte models. Front Microbiol 5:1–13. https://doi.org/10.3389/fmicb.2014.00776
Markovskaja S, Kačergius A, Davydenko K, Fraser S (2016) First record of Neocatenulostroma germanicum on pines in Lithuania and Ukraine and its co-occurrence with Dothistroma spp and other pathogens. For Pathol 46(5):522–533. https://doi.org/10.1111/efp.12308
McDougal R, Stewart A, Bradshaw R (2012) Transformation of Cyclaneusma minus with green fluorescent protein (GFP) to enable screening of fungi for biocontrol activity. Forests 3:83–94. https://doi.org/10.3390/f3010083
Merrill W, Wenner NG (1996) Cyclaneusma needle cast and needle retention in Scots pine. Plant Dis 80:294–298
Millar CS, Minter DW (1980) Naemacyclus minor. CMI Descriptions of Pathogenic Fungi and Bacteria. 66(659)
Minter DW, Dudka IO (1996) Fungi of Ukraine. A Preliminary Checklist. Egham, UK, International Mycological Institute and Kiev, Ukraine, M.G. Kholodny Institute of Botany, 361
Ondrušková E, Adamčík S, Kobza M, Jánošíková Z, Ostrovský R, Pastirčáková K, Caboň M, Adamčíková K (2023) Checking the balance between pathogenic and mutualistic pine needle fungi of the genus Lophodermium in forested and urban areas of Slovakia. Scand J For Res 38(1–2):39–48. https://doi.org/10.1080/02827581.2023.2191004
Pastirčáková K, Ivanová H, Pastirčák M (2014) Druhová diverzita húb na boroviciach (Pinus spp.) v mestskej a mimomestskej vegetácii. In: Zborník referátov z vedeckej konferencie “Dendrologické dni v Arboréte Mlyňany SAV 2014”. Arborétum Mlyňany SAV, pp 150–157
Prihatini I, Glen M, Wardlaw TJ, Mohammed CL (2014) Multigene phylogenetic study of Cyclaneusma species. For Path 44:299–309. https://doi.org/10.1111/efp.12101
Prihatini I, Glen M, Wardlaw TJ, Mohammed CL (2015) Diversity and identification of fungi associated with needles of Pinus radiata in Tasmania. South For J For Sci 78(1):19–34. https://doi.org/10.2989/20702620.2015.1092345
Sieber TN (1989) Endophytic fungi in twigs of healthy and diseased Norway spruce and white fir. Mycol Res 92:322–326. https://doi.org/10.1016/S0953-7562(89)80073-5
Sieber TN, Rys J, Holdenrieder O (1999) Mycobiota in symptomless needles of Pinus mugo ssp. uncinata. Mycol Res 103(3):306–310. https://doi.org/10.1017/S0953756298007229
Tarallo M, Dobbie K, Nunes Leite L, Waters T, Gillard K, Sen D, Mesarich CH, Bradshaw RE, McDougal RL (2023) Genomic, effector protein and culture-based analysis of Cyclaneusma minus in New Zealand provides evidence for multiple morphotypes. BioRxiv. https://doi.org/10.1101/2023.05.21.541640
Turis P, Valachovič M (2014) Secondary woody communities with Pinus nigra in Slovakia. Acta Carpatica Occidentalis 5:33–45
Watt MS, Palmer DJ, Bulman LS, Harrison D (2012a) Predicting the severity of Cyclaneusma needle cast on Pinus radiata under future climate in New Zealand. N Z J For Sci 42:65–71. https://doi.org/10.1139/x2012-021
Watt MS, Rolando CA, Palmer DJ, Bulman LS (2012b) Predicting the severity of Cyclaneusma minus on Pinus radiata under current climate in New Zealand. Can J For Res 42:667–674. https://doi.org/10.1139/x2012-021
White TJ, Bruns T, Lee S, Taylor JW (1990) PCR protocols: a guide to methods and applications: part three-genetics and evolution. In: Innis JA, Gelfand DH, Sninsky JJ, White TJ (eds) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Academic Press, London, pp 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Ying-Ren L (2012) Flora Fungorum Sinicorum: Rhytismatales, vol 40. Science Press, Beijing, p 261
Zúbrik M, Kunca A, Novotný J (2019) Hmyz a huby: Atlas poškodenia lesných drevín. Národné lesnícke centrum, Zvolen
Acknowledgements
The authors’ team wants to thank Mgr. Katarína Pastirčáková, PhD., the Plant Pathology Herbarium (NR) curator, for large sample collection processing, Ing. Peter Hoťka, PhD., an employee of the Mlyňany Arboretum for advice and guidelines on dendrological disputes, Monika Halandová and Mária Turčeková for technical work in the mycology laboratory and database processing.
Funding
Open access funding provided by The Ministry of Education, Science, Research and Sport of the Slovak Republic in cooperation with Centre for Scientific and Technical Information of the Slovak Republic. This work was supported by the Slovak Research Grant Agency VEGA project no. 2/0132/22 and by MVTS COST 20132. This work is based upon work from COST Action < CA20132—Urban Tree Guard—Safeguarding European urban trees and forests through improved biosecurity (UB3Guard) >, supported by COST (European Cooperation in Science and Technology).
Author information
Authors and Affiliations
Contributions
Emília Ondrušková presented investigation, conceptualization, methodology, data curation, formal analysis and writing—original draft preparation. Marek Kobza provided data curation and investigation. Zuzana Jánošíková performed investigation and founding acquisition. Rebecca McDougal carried out methodology, writing—reviewing and editing. Katarína Adamčíková analysed investigation, conceptualisation, writing—original draft preparation and supervision.
Corresponding author
Ethics declarations
Conflict of interest
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
41348_2024_924_MOESM1_ESM.pdf
Table S1 Details of Cyclaneusma samples from Slovakia, collected during 2014–2020 and analysed in this study using species specific primers according to Hunter et al. (2016) and Sanger sequencing (PDF 451 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ondruskova, E., Kobza, M., Janosikova, Z. et al. Which Cyclaneusma minus morphotypes are responsible for needle cast of Pinus spp. in Slovakia?. J Plant Dis Prot (2024). https://doi.org/10.1007/s41348-024-00924-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41348-024-00924-y