Abstract
Over the past decades, European ash trees in Germany have been affected by ash dieback, reducing their vigour and mechanical resistance. Those trees that also have stem collar necroses and the resulting stem rot are particularly affected. In this study, multilocus genotypes (MLGs) of Hymenoscyphus fraxineus and their interactions with other fungi from stem collar necroses were analysed. Ten ash trees from three different adjacent forest stands in central Germany were sampled. A total number of 716 isolates were obtained from stem collar necroses from these ten trees. Microsatellite analysis was successfully performed on 274 isolates identified as H. fraxineus and 26 MLGs were revealed. The number of MLGs varied from one to seven per tree and did not correspond to the number or severity of necroses. A striking result was that five of the MLGs occurred in two trees. All other MLGs occurred independently in only one tree, as expected. Our data show that when multiple MLGs were observed in a tree, one of the MLGs outnumbered the others, indicating that H. fraxineus is a primary coloniser of stem collar necroses. A total of 61 morphotypes, including H. fraxineus, were identified and discussed, comprising endophytic, saprotrophic and pathogenic fungi. Between five and 19 different fungi were found per stem collar necrosis. The majority of all isolated morphotypes were Ascomycota (82%), with the most common orders being Xylariales and Hypocreales. The most frequently isolated morphotypes, apart from H. fraxineus, were Armillaria sp. and Diplodia fraxini. Together they account for more than three quarters of all assigned isolations. Apart from H. fraxineus, only Diplodia fraxini was isolated from all ten trees.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The invasive fungus Hymenoscyphus fraxineus (T. Kowalski) Baral, Queloz & Hosoya (Helotiaceae, Ascomycota) is the cause of ash dieback, a severe disease that has affected European ash (Fraxinus excelsior L.; hereafter referred to as ash trees) since the early 1990s, initially in Poland and Lithuania, and is now widespread in Europe (Przybyl 2002; Lygis et al. 2005). The first confirmation of the pathogen in Germany was in 2006, but the disease has probably been present here since 2002 (Heydeck et al. 2005; Schumacher et al. 2007). Ash trees of all ages are infected with many different symptoms, including stem collar damage such as stem collar and root necrosis, or crown damage such as wilting, sunken cankers, leaf necrosis, inner bark discoloration, shoot blight or epicormic shoots (Gross et al. 2014a; Langer 2017). In terms of stem collar damage, stem collar necrosis (SCN) is a lesion on the stem consisting of necrotic tissue on the outside and inside. The form of the SCN is influenced by the fungi involved, the progression of the SCN according to the conditions present, or other factors. When the process is already more advanced, the initial infection by H. fraxineus is followed by other fungi that can cause wood rot at the base of the stem. The composition of these SCN associated fungal communities has recently been investigated in initial studies by Peters et al. (2023). As the fungus caused SCN spreads to the xylem and heartwood of ash trees, it reduces their stability and increases mortality (Langer 2017). Some of the fungi associated with SCN are able to decay wood. Wood-decaying fungi can be classified into three major groups based on their ability to degrade lignin and the type of wood decay: white rot fungi, brown rot fungi and soft rot fungi (Blanchette 1991; Langer et al. 2021). Through the production of ligninolytic extracellular oxidative enzymes, white rot fungi (Basidiomycota) are able to degrade lignin most effectively. The lignin removed from the wood reduces the stability of the tree and can cause the ash to fall (Linhares et al. 2021). A common white rot fungus on ash is, for example, Armillaria spp. (Sahu et al. 2021).
Even if the crown of an ash tree appears almost vital and healthy, SCN may be present on the trunk. However, they are more often found on severely damaged trees (Schumacher et al. 2010; Husson et al. 2012; Enderle et al. 2017; Langer 2017; Meyn et al. 2019). Hymenoscyphus fraxineus has a heterothallic mating system and produces asexual conidia in its anamorphic stage. The development and biology of the pathogen are described in detail in Gross et al. (2012b). Infection of ash by H. fraxineus is primarily caused by ascospores, which can be dispersed by wind over long distances. So far, there is no evidence that the asexually formed, sticky and non-infectious conidia of the anamorph play a significant role in the spread. They are thought to serve as spermatia during sexual reproduction (Gross et al. 2012b; Kirisits et al. 2013). However, there is also contrary information that H. fraxineus conidia may have an infectious effect (Fones et al. 2016).
Previous microsatellite analyses of H. fraxineus have already shown that there is a high genotypic diversity (Gross et al. 2014b; Nguyen et al. 2016) and that different multilocus genotypes (MLGs) can occur in ash shoots, branches and stems (Bengtsson et al. 2014; Meyn et al. 2019). Multiple infections by the pathogen are the most likely cause of different MLGs found in the same plant. There is no genetic differentiation between genotypes that infect the tree via the crown and those that attack at other compartments of the respective tree (e.g. at the stem base), and therefore all H. fraxineus MLGs seem to have the potential for stem collar infection (Burokiene et al. 2015).
This work is based on the hypothesis that one H. fraxineus individual per SCN indicates that the pathogen is a primary coloniser, biotrophically invading plant tissues. The presence of multiple H. fraxineus individuals per SCN would indicate a saprophytic lifestyle, with many individuals colonising pre-damaged tissue due to high spore pressure or a succession of multiple infections over time. This study is part of the FraxForFuture demonstration project and the FraxPath and FraxGen sub-networks (Langer et al. 2022). The aims of this research are to investigate the following questions: (i) How many H. fraxineus MLGs are involved in causing SCN in the ash trees studied? (ii) If the same H. fraxineus MLGs are present in SCN of different trees, are they different individuals? (iii) Is there a correlation between H. fraxineus MLGs and the type of SCN or other fungi associated with the SCN?
Materials and methods
Sampling site and stand
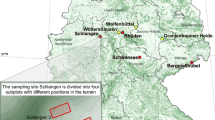
The sampling site selected for this study is not part of the common sampling site set in FraxForFuture (Langer et al. 2022). The three sampling plots are in close proximity (< 500 m distance) and are located in a mixed broadleaved forest site in Holzminden (Germany, Lower Saxony; hereafter referred to as forest site Holzminden; Fig. 1). The sampling plots have a comparable stand structure and a high proportion of ash trees. According to the forest management plan, and counting the annual rings of the sampled trees, these sampling plots are composed of trees that are approximately 35–45 years old. Plots I and II had a very similar proportion of ash (about 25%) and an ash tree density of 366 and 362 trees/ha, respectively. Plot III had a higher proportion (about 55%) and density (768 trees/ha) of ash trees. Other tree species mixed with F. excelsior were mainly Fagus sylvatica L., Acer pseudoplatanus L., Prunus avium L., Ulmus glabra Huds. and Tilia platyphyllos Scop. in decreasing proportions.
Location of the forest site Holzminden in Germany marked with an x (a). Consisting of three sampling plots and ten sampled trees. Crown condition of all ash trees in the sampling plots (b) and an enlarged view of sampling plots I, II and III with condition of the stem base (c-e). © GeoBasis-DE/BKG (2021); extract from the geodata of the Landesamt für Geoinformation und Landesvermessung Niedersachsen, © 2020 (QGIS Desktop 3.24.3)
The forest site Holzminden, with its three sampling plots, extends along a relatively steep (15–25°) western slope of a north–south oriented hill ridge in the southern part of Lower Saxony, within the Weser uplands. The slope is covered by a solifluction layer of Triassic limestone over claystone and sandstone of the upper bunter formation. Both substrates are nutrient-rich (calcareous-eutrophic), and well drained due to the steepness of the slope and the high coarse fraction in the solifluction cover layer. Therefore, the soil water regime of the plot I and plot II is relatively dry. Plot III is slightly moister than the other two plots due to its lower position on the slope and the higher proportion of clayey loam in the soil.
All ash trees on the plots were inspected and the stem base and the crown condition were classified following the guidelines of the ash dieback classification key (Peters et al. 2021a, 2021b) with slight modifications. The crown condition was rated on a scale of six damage classes (Fig. 1, in line with the mentioned key). The stem base was investigated slightly differently from the key, resulting in four damage classes depending on the proportion of affected surface.
Sampled trees
A total of ten ash trees (Online Resource 1) with a diameter at breast height between 10 and 16 cm were sampled in the forest site Holzminden. Trees 1, 2 and 3 were taken from sampling plot I, trees 4 and 5 from sampling plot II, and trees 6 to 10 from the southernmost sampling plot III (Fig. 1).
The ash trees were felled in March 2022 and cut at least 15 cm above the visible necrotic area at the stem base. The trunk bases and the uppermost parts of the main roots were then dug out with picks and shovels. Stem collars were transported to the laboratory in clean and labelled plastic bags.
Isolation and identification of fungi
In order to identify Dikarya Hibbett, T. Y. James & Vilgalys associated with SCN and to determine the fungal community, a culture-based method was chosen, including isolation of fungi from wood chips and identification of the resulting fungal species based on morphological observation and molecular analysis. The procedures for wood sample preparation, fungal culturing on malt yeast peptone (MYP; Langer 1994) agar medium, DNA extraction, PCR and sequencing of the ITS DNA region, subsequent morphotype (MT) designation and identification based on observed morphology and BLASTN results were carried out according to Peters et al. (2023). From each of the ten trees, three additional wood chips of asymptomatic stem tissue were taken as control samples. Fungi that could not be isolated because they were overgrown by other fungi or were contaminated, were grouped together as “Fungus sp.”. Yeasts and obvious plate contaminations were not considered. For each MT, at least one representative culture was placed in storage on MYP slant tubes at 4 °C in the fungal culture collection of the Northwest German Forest Research Institute (NW-FVA). Generated sequences were submitted to GenBank (Online Resource 2; http://www.ncbi.nlm.nih.gov/genbank; Altschul et al. 1997).
The frequency of each isolated fungal MT among all isolates was given as the percentage of that particular MT compared to all isolates. The continuity of isolated MTs was defined as the number of sampled trees in which the MT was detected in relation to the total number of sampled trees (Online Resource 2).
The relative abundances of the most common fungi relationships were evaluated visually using scatterplots. Relationships between these fungi and the number of MLGs were also evaluated. A possible relationship between the number of MLGs or the most common fungi and the fungal diversity was also analysed by comparing relative abundances. Trees were subject to varying sampling efforts with respect to fungal alpha diversity, making observed fungal alpha diversity numbers an outcome of a function of sample size (wood chips) per tree. Individual-based rarefaction (Gotelli and Colwell 2001) of samples from all other trees to the same number of samples is based on the principle: What fungal alpha diversity number would have been reported for each of the trees if all trees had the same sample size as the tree with the fewest samples? Rarefaction analysis (Sanders 1968) is methodologically based on a random subset of samples per tree and therefore needs to be repeated a sufficient number of times, here 1000, to fully exploit the tree-specific empirical sample. In each of the randomly generated sub-samples, the unique number of MTs is counted and divided by the total number of all unique MTs in all samples. The mean of these 1000 values is reported for each tree.
Microsatellite genotyping of Hymenoscyphus fraxineus
Microsatellite genotyping was performed on isolates that were morphologically identified as H. fraxineus. Fungal material was harvested by scraping fresh aerial mycelium from H. fraxineus cultures on agar plates. Homogenisation was performed in a Biolab Omni Bead Ruptor 24. (Biolab, Bebensee, Germany) for 2 × 30 s using five 1.4 mm ceramic beads (Omni International, Georgia, USA). H. fraxineus DNA was extracted using a Qiagen DNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The 20 microsatellite loci were amplified in a multiplex PCR, using the Qiagen Type-it ® Microsatellite PCR Kit (Qiagen) and fluorescence-labelled primers (Table 1). The primers were produced by biomers.net GmbH (biomers, Ulm, Germany). Fragments were amplified in a total reaction volume of 12.5 μl using 1 µl DNA template (5 ng/µl), 6.25 μl PCR multiplex mastermix, 2.75 μl distilled water 1.25 μl Q solution, and 1.25 μl primer mix (Table 1).
The PCR was performed in a Peqlab Thermocycler (Peqlab, Erlangen, Germany) with an initial denaturation step of 15 min followed by 30 cycles of 30 s denaturation at 95 °C, 90 s annealing at 56 °C and 60 s elongation at 72 °C. This was followed by a final elongation of 30 min at 60 °C. Fragment length analysis of the PCR product was performed in a GeXP DNA analyser (Beckman Coulter, Brea, USA). Fragment length determination and allele assignment were performed using the GeXP fragment analysis tool (Beckman Coulter). After checking each sample individually, an allele table was manually created in Excel.
Statistical analysis of microsatellite data
The Excel add-in GeneAlEx 6.2 was used to determine the number of different MLGs present in one sampled tree, considering each tree as a unique population. The probability PG that two or more individuals in a dataset are similar by chance was calculated for each MLG according to Milgroom (2015) (Online Resource 3), using the product of the allele frequencies for each respective locus.
Re-analysis of published crossing data
Published data from Gross et al. (2012b) was re-analysed to identify identical MLGs in the dataset of the crossing experiments. Gross et al. (2012b) were able to perform a cross between the strains Münc_r_41 and Münc_r_38 (Crossing A in Fig. 2) and a second cross between the strains Abts_22 and Abts_33 (Crossing B in Fig. 2), which resulted in apothecia with viable ascospores (Table 3 in Gross et al. 2012b). They prepared single-spore cultures from ascospores of the two crossings. In the original publication, allelic data from the offspring was used to confirm that the alleles of both parents were present in the 96 progeny single-spore cultures tested. The frequency of MLGs was not relevant in the study of Gross et al. (2012b).
Frequency of the occurrence of multilocus genotypes (MLGs) in the offspring of two crossings of Hymenoscyphus fraxineus individuals in a published crossing experiment (Gross et al. 2012b). Crossing A: strain Münc_r_38 × strain Münc_r_41, crossing B: strain Abts_22 × strain Abts_33
The aim of the re-analysis was to determine the potential of the microsatellite assay used in this study to differentiate closely related individuals. The similarity of the datasets (Online Resource 4) should allow conclusions to be drawn about the resolution of the primer assay of Gross et al. (2012b) and the one used in this study. The original allele table was kindly provided by Andrin Gross. The Excel add-in GenAlEx was used to determine the number of MLGs in the two full sibling populations.
Results
Sampling site and stand
A total of 703 ash trees were assessed to select ten sampled trees for analysis in this study. Almost 10% of all ash trees had already fallen, probably due to a combination of natural thinning and ash dieback. The rate of fallen ash trees was highest on plot III (with the highest proportion and density of ash) and lowest on plot II. Forty per cent of all ash trees had severe crown damage or were already dead (standing). More than half of these severely damaged ash trees were located on sampling plot III. Only five trees in all sampling plots showed very few symptoms of ash dieback in the crown. However, 41% of the trees, mainly located on sampling plot II, showed no damage at the base of the stem. The most severe SCN were mainly found on sampling plots I and III. Sampling plot III showed the most severe damage in the crown and at the base of the stem, followed by sampling plot I. Sampling plot II showed the least damage in the crown and at the base of the stem (Online Resource 5).
Sampled trees
The crown condition of the sampled trees ranged from slightly damaged to severely damaged (crown level 2 to 3 according to Peters et al. (2021a)). All trees had at least one obvious SCN with discoloured, sunken, and in some cases ruptured bark. The number of SCN per tree, based on the externally visible tops of the necrotic areas, varied from one to five. For seven out of ten sampled trees, more than half of the trunk was necrotic. Tree 6 had the lowest necrotic stem circumference with 25% of the stem being necrotic, while tree 7 had the highest with 90%. The ratio of the height of the examined SCN to the diameter of the stem base was on average two and a half times the diameter of the stem base. In four out of ten cases, a ruptured wood surface was visible at the SCN. With the exception of tree 1, white rot was visible in all trees (Table 2).
Isolated fungi
A total of 1497 wood chips of stem collar tissue originating from ten ash trees were incubated. Overall, 716 mycelial outgrowths were observed on 676 wood chips (45% of all chips incubated). Thirty outgrowths were unidentifiable due to contamination and were classified as “Fungus sp.”, giving a total of 686 fungal strains. Altogether, 377 chips (25%) showed no outgrowth at all after four weeks of incubation, while 278 (19%) chips had been overgrown by fast-growing fungi from adjacent wood chips before outgrowth could be detected. The remaining 166 (11%) wood chips were colonised or contaminated by yeasts, moulds, Trichoderma spp. or non-dikarya fungi. The resulting 686 fungal strains were assigned to 61 MTs (excluding Trichoderma spp.), comprising endophytic, saprotrophic and pathogenic fungi. Thirty-two MTs could be identified to species level. Twenty-two isolates could be identified to at least genus level (Online Resource 2). The majority of all isolated MTs were Ascomycota (50 MTs, 82%), 11 MTs (18%) belonged to the division of Basidiomycota. Within the Ascomycota, the most common orders were Xylariales and Hypocreales (both 22%), followed by Pleosporales (14%), and Helotiales (12%). The Basidiomycota fungi were mainly represented by Russulales, Agaricales and Polyporales (three species each).
Despite the diversity of the 61 fungal MTs detected, only five fungi had a frequency of more than 2%. The most frequent isolated MTs (Online Resource 2) were H. fraxineus (40.5%), followed by Armillaria sp. (22%), Diplodia fraxini (Fr.) Fr. (15.2%), Diaporthe cf. eres (3.1%) and Paracucurbitaria sp. (2.5%). Diplodia fraxini and H. fraxineus were the most continuous MTs, occurring on all studied trees (100%), followed by Armillaria sp. (90%). Together, the latter three MTs already accounted for more than three quarters of all assigned isolations. In at least half of the trees, the MTs Paracucurbitaria sp., Fusarium cf. lateritium and Angustimassarina sp. were also present. Furthermore, the MTs Angustimassarina sp., Armillaria sp., Diaporthe cf. eres, D. fraxini, Fusarium cf. lateritium, H. fraxineus and Paracucurbitaria sp. were isolated from all three sampling plots.
Between five and 19 different fungi, including H. fraxineus, were found per SCN. On average, 12 MTs were recorded per tree. Forty-two of the 61 MTs were isolated from a single tree. The MTs Cytospora sp., Hypocreales sp., and Vuilleminia sp. were only isolated from asymptomatic tissue.
The graphical comparison of the relative abundances of the most common fungi Armillaria spp., D. fraxini and H. fraxineus with each other and with the MLG numbers, as well as the most common fungi and the MLG numbers with the fungal diversity showed no clear evidence of underlying strong dependencies (figures not shown).
Population genetics of Hymenoscyphus fraxineus
Genetic diversity
The multiplex primers identified 26 different MLGs (A-Z) in the set of 274 H. fraxineus samples derived from ten sampled trees with different numbers of SCN (Online Resource 6), indicating a low genotypic diversity. The loci Chafra02 and mHp_077098 were monomorphic and therefore excluded from further analysis. Allelic diversity was low, with the majority of loci (16/18) being biallelic. The locus mHp_088853 showed a third allele in two isolations from sampled tree 9. The locus Chafra03 had a maximum of five alleles, with the most frequent alleles being 210 and 190. The other three alleles occurred exclusively in sampled trees 6 and 7. While the alleles 212 and 214 occurred with a high frequency, allele 229 occurred only once.
Nine H. fraxineus MLGs were found in sampling plot I, two in sampling plot II and 15 in sampling plot III. Considering the different number of sampled trees in each sampling plot, the proportion of MLGs per sampling plot was three (I), one (II) and three (III). The largest number of H. fraxineus MLGs (7) was found in sampling plot III, where the highest number of samples was taken because the crown and stem collar damages due to ash dieback (Fig. 1) were most severe.
Probability of identity
To determine the probability of two or more individuals in a dataset being similar by chance, the probability of identity PG was calculated. The calculated PG values ranged from 10−4 to 10−7 (Online Resource 3), which suggests that in the considered dataset of 274 samples it is unlikely to find two MLGs that are identical by chance, assuming that the analysed individuals are unrelated.
Re-analysis of published data
To assess the resolution power of the primer assay used in this study, published data from Gross et al. (2012b) of two crossing experiments of single-spore cultures was re-analysed. The majority (12/20) of the primers used in this study were also used in Gross et al. (2012b) (Online Resource 4), allowing conclusions to be drawn about the resolution power of the primer assay. Furthermore, the total number of primers used in both studies was similar.
Due to some missing data in the original allelic tables of 2 × 96 single-spore cultures, re-analysis was possible for 88 (crossing A) and 95 (crossing B) single-spore cultures. The re-analysis showed the occurrence of identical MLGs within both of the crossing experiments (Fig. 2). Among the 88 descendants of the Münc_r_41 × Münc_r_38 crossing, only eight unique MLGs could be identified. Every other MLG occurred at least twice and a maximum of nine times. In the Abts_22 × Abts_33 crossing, 38 of the 95 descendants had unique MLGs. This suggests that a set of 19 primers representing 17 nuclear polymorphic loci cannot reliably distinguish between full sibling individuals. Therefore, the microsatellite array used in this study, with 20 primers representing 18 nuclear polymorphic loci, is also unlikely to reliably distinguish between closely related individuals.
Number of multilocus genotypes per sampled tree
In order to assess the genetic diversity within a tree, the number of individuals per sampled tree was determined, whereas each individual is represented by one MLG. In sampled trees 4, 5 and 8, only a single MLG was present. In trees with multiple MLGs, one MLG outnumbered the others with a frequency of more than 50% (Fig. 3, Online Resource 6). This was the case for all sampled trees except 7 and 9. A maximum of seven MLGs could be identified in sampled tree 7, without one MLG dominating the others (Fig. 3). This was also the tree with the largest number of samples.
Five (L, M, N, O and P) of the seven MLGs occurring in sampled tree 7 were also isolated from sampled tree 6. These two trees grew relatively close to each other compared to the possible pairwise distances of all sampled trees analysed (Fig. 4).
The frequency of occurrence of the identical MLGs in sampled tree 6 and 7 was different in both trees (Fig. 3). While L was the most frequently isolated MLG in sampled tree 6, sampled tree 7 was dominated by MLG O. Some of the shared MLGs (M, N and O) between sampled tree 6 and sampled tree 7 had alleles (212, 214) at locus Chafra03 that are unique in the dataset (Online Resource 6).
Number of multilocus genotypes per stem collar necrosis
The aim of this study was to determine the number of H. fraxineus MLGs present in a single SCN. As the SCN of trees with multiple infections tended to overlap, a distinct spatial assignment was often not possible. In no tree, except for sampled tree 10, did the number of detected SCN correspond to the number of MLGs present (Table 2, Online Resource 6). Sampled tree 7 had the highest number of MLGs (7) and at the same time the most visible SCN (5), but it was also evaluated with the second highest number of incubated wood chips (Table 2, Fig. 3, Online Resource 6).
Furthermore, the number of MLGs seems to be independent of the size of the SCN, as both the lowest number of MLGs (1) and the highest number (7) were found on trees with large, advanced SCN (Table 2).
As sampled trees 6 and 7 were the only trees with identical MLGs, the focus lies on these two sampled trees. In sampled tree 6, two SCN were identified, based on the externally visible top ends of the necrotic areas. For the majority of both SCN, L was isolated with the highest frequency. The other common MLGs, including M, N, O and P, were only found at the top end of the SCN (Fig. 5).
Multilocus genotypes (MLGs) in sampled tree 6. Cuttings (dashed lines) and corresponding sections (arrow) are shown as red lines. The extent of the stem collar necroses from the outside is represented by the blue transparent area. The location of the different MLGs is indicated with corresponding colours and letters. Externally visible upper ends of necrotic areas are marked with white asterisks. In total, there were 27 Hymenoscyphus fraxineus isolation sites with five different MLGs
Sampled tree 7 (Fig. 6) showed the highest number of SCN as well as the highest number of MLGs. L, M, N, O and P MLGs were also present in sampled tree 7, while M and O were isolated with almost equal frequency. The distribution of MLGs in the stem collar of sampled trees 6 and 7 resembled a spatial clustering structure, because each SCN top end was dominated by a distinct MLG.
Multilocus genotypes (MLGs) in sampled tree 7. Cuttings (dashed lines) and corresponding sections (arrow) are shown as red lines. The extent of the stem collar necroses from the outside is represented by the blue transparent area. The necrotic tissue at the back and the side of the stem are shown separately in their respective perspectives with blue arrow lines. The location of the different MLGs is indicated with corresponding colours and letters. Externally visible upper ends of necrotic areas are marked with white asterisks. In total, there were 55 Hymenoscyphus fraxineus isolation sites with seven different MLGs
Discussion
Identical multilocus genotypes in different sampled trees
One aim of this study was to determine the number of H. fraxineus individuals per SCN. In this context, it is important that the discriminatory power of the microsatellite assay used is high enough to distinguish individual MLGs. Resolution depends on the number of polymorphic loci analysed as well as on the number of alleles per locus. Most population genetic studies of H. fraxineus have used between 11 (Burokiene et al. 2015; Haňáčková et al. 2017) and 19 (Gross et al. 2012b; Meyn et al. 2019) microsatellite loci. While Meyn et al. (2019) were able to resolve all analysed samples, Gross et al. (2012b), Burokiene et al. (2015), and Haňáčková et al. (2017) reported identical MLGs. The trees analysed in Meyn et al. (2019) originated from a contiguous sampling site, where the maximum distance between analysed trees was 29 m. However, the total sample size of four trees was relatively small, decreasing the chance of finding identical MLGs.
There are several explanations to find identical MLGs. Asexual reproduction would lead to prevalence of a low number of genotypes at the population level. All available population genetics studies (e.g. Gross et al. 2012b, 2014b; Haňáčková et al. 2015, 2017; Burokiene et al. 2015) show a high genetic diversity at the population level supporting the current model of H. fraxineus reproducing exclusively sexually via ascospores (Gross et al. 2014a). In the present study, the presence of the same MLG in a stem collar necrosis of a single tree (Figs. 5, 6) is interpreted as mycelial growth originating from a single ascospore. However, it remains to be explained why the same MLGs are found on trees 6 and 7, because they cannot present a single contiguous mycelium. A possible explanation is the occurrence of twins during ascospore development. An ascus is dikaryotic, containing one nucleus from the male and one from the female reproductive structure. These two nuclei fuse via karyogamy to form a diploid nucleus. Subsequent meiosis produces four haploid nuclei and is followed by mitosis, in which each nucleus divides again (Landvik et al. 2003). Thus, there are four times two identical spores in one ascus. Sampled trees 6 and 7 have a small distance between them (27 m) compared to the other pairwise tree distances (Fig. 4). Nevertheless, it is highly unlikely that two genetically identical spores would fall on these two trees.
The sampled trees with identical MLGs are located in sampling plot III with the highest density of ash trees and the most damaged crowns and stem collars (Online Resource 5). At this sampling plot, the pathogen population is the largest, which increases the probability of finding identical MLGs. H. fraxineus individuals from European populations exhibit predominantly biallelic loci (Gross et al. 2014b), implying a lower discriminatory power of the loci used, which increases the difficulty to distinguish between individuals. This is particularly severe when the individuals studied are closely related. The re-analysed data of Gross et al. (2012b) showed that in a full sibling population not all individuals can be fully resolved using a primer assay of 12 nuclear polymorphic markers. Therefore, the authors suggest that the identical MLGs in sampled trees 6 and 7 are closely related individuals and thus provide an indication of the answer to the question (ii) whether the same H. fraxineus MLGs in the SCN of different trees are different individuals. A higher genetic resolution could only be achieved with other methods, for instance, population genetics with whole genomes. A more traditional and morphology-based approach to determine whether the H. fraxineus individuals in sample trees 6 and 7 are very closely related or the same individual would be a mating test with mycelia of the different isolates in dual cultures. Such mating tests, not presented in this study, have been carried out, but due to the slow growth of H. fraxineus and the often highly variable culture habitus of a single strain, no reliable results could be obtained.
Colonisation patterns in stem collar necrosis
Another aim of this study was to determine whether the number of H. fraxineus individuals in a SCN would allow to differentiate between primary and secondary infections. The dominance of one MLG would be considered as a sign of primary infection causing the SCN, whereas an equal distribution of several MLGs would indicate secondary infection of an existing SCN. A tendency for one MLG to outnumber others within a tree and a spatial clustering structure of MLGs within a SCN were found in this study. This pattern is similar to the infection pattern found in ash shoots, where successful shoot infections are characterised by the dominance of one MLG (Haňáčková et al. 2017). Because H. fraxineus is the primary agent of ash dieback, which initially infects the shoot, the observed pattern is consistent with other studies suggesting H. fraxineus plays also a causal role in SCN emergence (Husson et al. 2012; Chandelier et al. 2016; Langer 2017). If H. fraxineus was a secondary coloniser, a more random distribution of MLG frequencies would be expected. Instead, the infection pattern probably established successively over time, assuming that all MLGs found in a SCN are successful pathogens, and the different MLGs probably represent multiple, temporally staggered infections. It is likely that the first MLG that successfully invades the stem become dominant because it has the temporal lead, which is then reflected in the size of the SCN.
Associated fungi and the cohabitation role of Hymenoscyphus fraxineus
As a continuation of the study by Peters et al. (2023), 23 additional MTs were found at the forest site Holzminden. Of these, three species, Lopadostoma turgidum (Pers.) Traverso, Stachybotrys chlorohalonatus B. Andersen & Thrane, and Trimmatostroma betulinum (Corda) S. Hughes, are described on ash for the first time. The detection of L. turgidum in woody tissues of ash is noteworthy, as Jaklitsch et al. (2014) could only find species of the genus Lopadostoma (Nitschke) Traverso in the bark of trees and shrubs, but never in underlying wood. The stromata of L. turgidum usually occur on F. sylvatica, which is one of the tree species present in the forest site Holzminden. (Chapter 2.1). Stachybotrys chlorohalonatus (Stachybotryaceae, Hypocreales) has been separated from S. chartarum (Ehrenb.) S. Hughes sensu lato as a distinct species, which differs morphologically and in the tri5, chs1 and tub1 gene fragment sequences (Andersen et al. 2003). This black mould was isolated from a cardboard on gypsum board in Denmark, a wet cellulose-containing material, the typical habitat of the species. Trimmatostroma betulinum (Mollisiaceae, Helotiales) is known to be a naturally occuring member of the fungal community that inhabits living leaves and twigs of Betula spp. without causing symptoms, but can also been found on living or dead twigs, branches and occasionally leaves of other plants including Pinus, Alnus, Quercus, and Salix species (UK, CAB International et al. 2003).
The 23 additional MTs represent approximately one-third of all fungi isolated (61) from the sampled SCN. This result confirms the hypothesis of Peters et al. (2023) that the number of associated species may increase as more different forest plots are examined. As expected, the most common fungi in this study (Armillaria sp., Diaporthe cf. eres, D. fraxini, H. fraxineus, and Paracucurbitaria sp.) match with the most common fungi isolated by Peters et al. (2023) and other previous studies on fungi associated with ash necroses (Lygis et al. 2005; Langer 2017; Linaldeddu et al. 2020). In contrast to the results of Langer (2017), Meyn et al. (2019), and Peters et al. (2023), the frequency and continuity of Neonectria punicea (J. C. Schmidt) Castl. & Rossman, represented by only two isolates, was significantly lower in this study. However, similar to the results of Peters et al. (2023), at least a quarter of all MTs were identified as being able to decay wood (Online Resource 2).
The 274 H. fraxineus isolates and the occurrence and composition of the resulting 25 MLGs did not seem to be influenced by the presence of other isolated species or fungal diversity in any of the ten sampled trees. In general, a correlation would be conceivable, as H. fraxineus is a slow-growing fungus that could be overgrown by fast-growing fungi after their secondary colonisation (Kowalski and Holdenrieder 2009; Gross et al. 2014a). However, our results showed that neither the number of H. fraxineus isolates nor the number of MLGs were lower when the most common species, Armillaria spp. or D. fraxini, were present. The analysis showed that the fungal diversity (number of species) in SCN did not influence the composition of H. fraxineus or MLGs. This trend is unlikely to change, although theoretically more infections are possible, but individual strains may not be isolated from the wood.
The 61 MTs isolated included various endophytes, saprotrophs and pathogens. In this study, the most consistently detected isolates, besides H. fraxineus, belonged to the genus Armillaria (Fr.) Staude or could be identified as D. fraxini. It is likely that species of the genus Armillaria were present in European forests prior to ash dieback, as they are known to be widespread soil colonisers (Morrison 2004; Lygis et al. 2005; Bakys et al. 2009). Their common occurrence in advanced SCN can be explained by their ascribed property of being secondary pathogens that attack stressed trees and are able to decay wood (Chandelier et al. 2016). Armillaria spp. can colonise stem collars after SCN has been caused by H. fraxineus, just as Armillaria spp. can independently infect a weakened tree without SCN caused by H. fraxineus (Langer 2017). Irrespective of the individual infection process, this leads to an accelerated disease in ash trees affected by ash dieback. But even in the latter case, Armillaria spp. is only a secondary coloniser, because the tree has already been pre-damaged in the crown by the pathogen that causes ash dieback. Diplodia fraxini, on the other hand, is known to be an aggressive pathogen, causing bark lesions and wood discoloration or to enlarge necroses, which are primarily caused by H. fraxineus (Alves et al. 2014; Linaldeddu et al. 2020, 2022). Kowalski et al. (2017) ranked D. fraxini as the second most pathogenic fungus after H. fraxineus, although it was not mentioned as a frequent coloniser of F. excelsior prior to ash dieback (Kowalski et al. 2016). Diplodia fraxini undoubtedly contributes to a greater extent of damaged stem collars and therefore plays an important role in the development of this disease.
Other, less common fungi, such as Neonectria punicea, can occur as latent pathogens, meaning that they are already present in the tree as endophytes prior to damage and change their life cycle to become pathogenic when the host is weakened (Langer 2017). Such fungal characteristics may also increase SCN development by weakening the tree in the context of ash dieback.
Conclusion and outlook
The study shows that in most sampled trees one MLG outnumbers the others, indicating that a SCN was initiated by a single H. fraxineus MLG. It is therefore suggested that H. fraxineus is a primary coloniser of SCN. The following conclusions can be drawn:
-
1.
Because of the close proximity, it is reasonable to assume that identical MLGs in a SCN result from growth of a single individual rather than multiple infections.
-
2.
Identical MLGs in different trees could result from infections of closely related individuals.
-
3.
Multiple MLGs within one tree may represent multiple successive infections over time.
In this study, the authors have shown the limitations of using microsatellites to resolve individuals. In future population genetic studies, other molecular markers that cover larger genome sections should be used, especially when close genetic relationships are suspected. One method could be single-nucleotide polymorphism analysis based on genome sequencing (population genomics).
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein databases search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Alves A, Linaldeddu BT, Deidda A, Scanu B, Phillips AJL (2014) The complex of Diplodia species associated with Fraxinus and some other woody hosts in Italy and Portugal. Fungal Divers 67:143–156. https://doi.org/10.1007/s13225-014-0282-9
Andersen B, Nielsen KF, Thrane U, Szaro T, Taylor JW, Jarvis BB (2003) Molecular and phenotypic descriptions of Stachybotrys chlorohalonata sp. nov. and two chemotypes of Stachybotrys chartarum found in water-damaged buildings. Mycologia 95:1227–1238. https://doi.org/10.1080/15572536.2004.11833031
Bakys R, Vasaitis R, Barklund P, Thomsen IM, Stenlid J (2009) Occurrence and pathogenicity of fungi in necrotic and non-symptomatic shoots of declining common ash (Fraxinus excelsior) in Sweden. Eur J for Res 128:51–60. https://doi.org/10.1007/s10342-008-0238-2
Bengtsson SBK, Vasaitis R, Kirisits T, Solheim H, Stenlid J (2012) Population structure of Hymenoscyphus pseudoalbidus and its genetic relationship to Hymenoscyphus albidus. Fungal Ecol 5:147–153. https://doi.org/10.1016/j.funeco.2011.10.004
Bengtsson SBK, Barklund P, von Brömssen C, Stenlid J (2014) Seasonal pattern of lesion development in diseased Fraxinus excelsior Infected by Hymenoscyphus pseudoalbidus. PLoS ONE 9(4):e76429. https://doi.org/10.1371/journal.pone.0076429
Blanchette R (1991) Delignification by wood-decay fungi. Annu Rev Phytopathol 29:381–403. https://doi.org/10.1146/annurev.py.29.090191.002121
Burokiene D, Prospero S, Jung E, Marčiulynienė D, Moosbrugger K, Norkute G, Rigling D, Lygis V, Schoebel C (2015) Genetic population structure of the invasive ash dieback pathogen Hymenoscyphus fraxineus in its expanding range. Biol Invasions 17:2743–2756. https://doi.org/10.1007/s10530-015-0911-6
Chandelier A, Gerarts F, Martin GS, Herman M, Delahaye L (2016) Temporal evolution of collar lesions associated with ash dieback and the occurrence of Armillaria in Belgian forests. For Pathol 46:289–297. https://doi.org/10.1111/efp.12258
Enderle R, Sander F, Metzler B (2017) Temporal development of collar necroses and butt rot in association with ash dieback. iForest 10:529–536. https://doi.org/10.3832/ifor2407-010
Fones HN, Mardon C, Gurr SJ (2016) A role for the asexual spores in infection of Fraxinus excelsior by the ash-dieback fungus Hymenoscyphus fraxineus. Sci Rep 6(1):34638. https://doi.org/10.1038/srep34638
Gherghel F, Fussi B, Donges K, Haustein M, Jakob KM, Müller K, Piškur B, Hauptman T, Lenz HD, Konnert M, Kost G, Rexer KH (2014) The development of a species-specific test to detect Hymenoschyphus pseudoalbidus in ash tissues. For Pathol 44:137–144. https://doi.org/10.1111/efp.12078
Gotelli NJ, Colwell RK (2001) Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett 4:379–391. https://doi.org/10.1046/j.1461-0248.2001.00230.x
Gross A, Grünig CR, Queloz V, Holdenrieder O (2012a) A molecular toolkit for population genetic investigations of the ash dieback pathogen Hymenoscyphus pseudoalbidus. For Pathol 42:252–264. https://doi.org/10.1111/j.1439-0329.2011.00751.x
Gross A, Zaffarano PL, Duo A, Grünig CR (2012b) Reproductive mode and life cycle of the ash dieback pathogen Hymenoscyphus pseudoalbidus. Fungal Genet Biol 49:977–986. https://doi.org/10.1016/j.fgb.2012.08.008
Gross A, Holdenrieder O, Pautasso M, Queloz V, Sieber TN (2014a) Hymenoscyphus pseudoalbidus, the causal agent of European ash dieback. Mol Plant Pathol 15:5–21. https://doi.org/10.1111/mpp.12073
Gross A, Hosoya T, Queloz V (2014b) Population structure of the invasive forest pathogen Hymenoscyphus pseudoalbidus. Mol Ecol 23:2943–2960. https://doi.org/10.1111/mec.12792
Haňáčková Z, Koukol O, Havrdová L, Gross A (2015) Local population structure of Hymenoscyphus fraxineus surveyed by an enlarged set of microsatellite markers. For Pathol 45(5):400–407. https://doi.org/10.1111/efp.12185
Haňáčková Z, Koukol O, Čmoková A, Zahradník D, Havrdová L (2017) Direct evidence of Hymenoscyphus fraxineus infection pathway through the petiole-shoot junction. For Pathol 47:e12370. https://doi.org/10.1111/efp.12370
Heydeck P, Bemmann M, Kontzog HG (2005) Triebsterben an Gemeiner Esche (Fraxinus excelsior) im nordostdeutschen Tiefland. Forst Und Holz 60:505–506
Husson C, Caël O, Grandjean JP, Nageleisen LM, Marçais B (2012) Occurrence of Hymenoscyphus pseudoalbidus on infected ash logs. Plant Pathol 61:889–895. https://doi.org/10.1111/j.1365-3059.2011.02578.x
Jaklitsch WM, Fournier J, Rogers JD, Voglmayr H (2014) Phylogenetic and taxonomic revision of Lopadostoma. Persoonia 32:52–82. https://doi.org/10.3767/003158514X679272
Kirisits T, Dämpfle L, Kräutler K (2013) Hymenoscyphus albidus is not associated with an anamorphic stage and displays slower growth than Hymenoscyphus pseudoalbidus on agar media. For Pathol :n/a-n/a. https://doi.org/10.1111/efp.12042
Kowalski T, Holdenrieder O (2009) Pathogenicity of Chalara fraxinea. For Pathol 39:1–7. https://doi.org/10.1111/j.1439-0329.2008.00565.x
Kowalski T, Kraj W, Bednarz B (2016) Fungi on stems and twigs in initial and advanced stages of dieback of European ash (Fraxinus excelsior) in Poland. Eur J for Res 135:565–579. https://doi.org/10.1007/s10342-016-0955-x
Kowalski T, Bilański P, Kraj W (2017) Pathogenicity of fungi associated with ash dieback towards Fraxinus excelsior. Plant Pathol 66:1228–1238. https://doi.org/10.1111/ppa.12667
Landvik S, Schumacher TK, Eriksson OE, Moss ST (2003) Morphology and ultrastructure of Neolecta species. Mycol Res 107:1021–1031. https://doi.org/10.1017/S0953756203008219
Langer GJ (2017) Collar rots in forests of Northwest Germany affected by ash dieback. In: Enderle R, Pliura A, Vaisatis R (eds) Dieback of European Ash (Fraxinus spp.)—Consequences and guidelines for sustainable management. SLU Uppsala, Uppsala, pp 4–19
Langer GJ, Bußkamp J, Blumenstein K, Terhonen E (2021) Fungi inhabiting woody tree tissues—stems, branches, and twigs. Forest Microbiome. Elsevier, London, pp 175–205
Langer GJ, Fuchs S, Osewold J, Peters S, Schrewe F, Ridley M, Kätzel R, Bubner B, Grüner J (2022) FraxForFuture—research on European ash dieback in Germany. J Plant Dis Prot 129:1285–1295. https://doi.org/10.1007/s41348-022-00670-z
Langer GJ (1994) Die Gattung Botryobasidium DONK (Corticiaceae, Basidiomycetes). J. Cramer, Berlin, Stuttgart
Linaldeddu BT, Bottecchia F, Bregant C, Maddau L, Montecchio L (2020) Diplodia fraxini and Diplodia subglobosa: the main species associated with cankers and dieback of Fraxinus excelsior in North-Eastern Italy. Forests 11:883. https://doi.org/10.3390/f11080883
Linaldeddu BT, Bregant C, Montecchio L, Brglez A, Piškur B, Ogris N (2022) First report of Diplodia fraxini and Diplodia subglobosa causing canker and dieback of Fraxinus excelsior in Slovenia. Plant Dis 106:26–29. https://doi.org/10.1094/PDIS-06-21-1204-SC
Linhares CSF, Gonçalves R, Martins LM, Knapic S (2021) Structural stability of urban trees using visual and instrumental techniques: a review. Forests 12(12):1752. https://doi.org/10.3390/f12121752
Lygis V, Vasiliauskas R, Larsson KH, Stenlid J (2005) Wood-inhabiting fungi in stems of Fraxinus excelsior in declining ash stands of northern Lithuania, with particular reference to Armillaria cepistipes. Scand J for Res 20:337–346. https://doi.org/10.1080/02827580510036238
Meyn R, Langer GJ, Gross A, Langer EJ (2019) Fungal colonization patterns in necrotic rootstocks and stem bases of dieback-affected Fraxinus excelsior L. For Pathol 49:e12520. https://doi.org/10.1111/efp.12520
Milgroom MG (2015) Population biology of plant pathogens: genetics, ecology, and evolution. The American Phytopathological Society, Minnesota
Morrison DJ (2004) Rhizomorph growth habit, saprophytic ability and virulence of 15 Armillaria species. For Pathol 34:15–26. https://doi.org/10.1046/j.1439-0329.2003.00345.x
Nguyen DT, Cleary MR, Enderle R, Berlin A, Stenlid J (2016) Analyses of the ash dieback pathogen, Hymenoscyphus fraxineus, suggest role of tree species diversity on colonization and population structure differentiation. For Pathol 46:82–84. https://doi.org/10.1111/efp.12236
Peters S, Langer G, Kätzel R (2021b) Bonitur geschädigter Eschen im Kontext des Eschentriebsterbens. AFZ - DerWald 76:28–31
Peters S, Fuchs S, Bien S, Bußkamp J, Langer GJ, Langer EJ (2023) Fungi associated with stem collar necroses of Fraxinus excelsior affected by ash dieback. Mycol Prog 22:52. https://doi.org/10.1007/s11557-023-01897-2
Peters S, Langer G, Kätzel R (2021a) Eschentriebsterben - Kriterien zur Schadensbonitur an Eschen. Fachagentur für nachwachsende Rohstoffe e.V. (FNR), Gülzow-Prüzen
Przybyl K (2002) Fungi associated with necrotic apical parts of Fraxinus excelsior shoots. For Pathol 32(6):387–394. https://doi.org/10.1046/j.1439-0329.2002.00301.x
Sahu N, Merényi Z, Bálint B, Kiss B, Sipos G, Owens RA, Nagy LG (2021) Hallmarks of Basidiomycete Soft- and White-Rot in Wood-Decay -Omics Data of Two Armillaria Species. Microorganisms 9(1):149. https://doi.org/10.3390/microorganisms9010149
Sanders HL (1968) Marine Benthic Diversity: A Comparative Study. Am Nat 102:243–282. https://doi.org/10.1086/282541
Schumacher J, Wulf A, Leonhard S (2007) Erster Nachweis von Chalara fraxinea T. KOWALSKI sp. nov. in Deutschland – ein Verursacher neuartiger Schäden an Eschen. Nachrichtenblatt Dtsch Pflanzenschutzdienstes 59:121–123
Schumacher J, Kehr R, Leonhard S (2010) Mycological and histological investigations of Fraxinus excelsior nursery saplings naturally infected by Chalara fraxinea. For Pathol 40:419–429. https://doi.org/10.1111/j.1439-0329.2009.00615.x
UK, Cab International, Bogomolova EV, Minter DW (2003) Trimmatostroma betulinum. Descr Fungi Bact 156:1560. https://doi.org/10.1079/DFB/20056401560
Acknowledgements
The authors would like to thank Peter Gawehn, Annette Ihlemann, Martina Hille, Daniel Gaunitz and Dr. Holger Sennhenn-Reulen for their technical and statistical support. The authors are also grateful to the Neuhaus Forestry Department for permission to sample the trees. Many thanks to Dr. Andrin Gross for generously providing his raw data for re-analysis. Special thanks to Maia Ridley for proof-reading the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. The project receives funding via the Waldklimafonds (WKF) funded by the German Federal Ministry of Food and Agriculture (BMEL) and Federal Ministry for the Environment, Nature Conservation, Nuclear Safety and Consumer Protection (BMUV) administrated by the Agency for Renewable Resources (FNR) under Grant Agreement Nos. 2219WK21A4, 2219WK22A4, and 2219WK22D4.

Author information
Authors and Affiliations
Contributions
Conceptualisation, Ben Bubner, Gitta Jutta Langer, and Sandra Peters; Funding acquisition, Gitta Jutta Langer and Ben Bubner; Investigation, Sandra Peters, Steffen Bien, and Nina Gruschwitz; Methodology, Viktoria Blunk, Steffen Bien, and Ben Bubner; Project administration, Gitta Jutta Langer and Ben Bubner; Supervision, Gitta Jutta Langer, Ben Bubner, and Ewald Johannes Langer; Writing—original draft, Sandra Peters and Nina Gruschwitz; Writing—commenting & editing, Viktoria Blunk, Steffen Bien, Sebastian Fuchs, Gitta Jutta Langer, Ben Bubner and Ewald Johannes Langer.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest. There are only non-financial research interests, related directly or indirectly to the work submitted for publication here.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peters, S., Gruschwitz, N., Bien, S. et al. The fungal predominance in stem collar necroses of Fraxinus excelsior: a study on Hymenoscyphus fraxineus multilocus genotypes. J Plant Dis Prot (2024). https://doi.org/10.1007/s41348-024-00912-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41348-024-00912-2