Abstract
The reaction of 636 Solanum (sections Lycopersicon and Juglandifolia) accessions were evaluated under greenhouse conditions after mechanical inoculation with a Jordanian isolate of the new tobamovirus tomato brown rugose fruit virus (ToBRFV). Local and systemic infections were assayed by symptoms evaluation and virus detection via biotests and RT-PCR. All cultivated tomatoes (Solanum lycopersicum) and the great majority of wild tomato accessions proved susceptible to ToBRFV. They showed a wide range of symptoms (mosaic, leaf deformations, mottling, shoestring, and stunting). Twenty-six accessions representing S. lycopersicum var. cerasiforme, S. pimpinellifolium, S. habrochaites, and S. chilense were tolerant. High levels of resistance have been demonstrated in three accessions of S. ochrantum, a close relative to wild tomatoes (member of the sect. Juglandifolia) not only to ToBRFV but also to the tobamoviruses, tobacco mosaic virus (TMV) and tomato mosaic virus (ToMV). After mechanical inoculation, the three tobamoviruses could be detected only in inoculated leaves in the accessions LA2160, LA2162, and LA 2166, which remained symptomless. However, two other S. ochrantum accessions PI 473,498 and PI 230,519 reacted unusually. They were demonstrated highly resistant to TMV and ToMV, but proved transiently susceptible to ToBRFV showing mild systemic mosaic followed by total recovery from symptoms and the virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tomato (Solanum lycopersicum) is one of the most important vegetables produced and consumed worldwide (Heuvelink 2018). Similar to other solanaceous plants, pathogens often cause heavy losses of fruit quality and yield of the crop. Tomato is susceptible to a wide range of viruses, out of which tobacco mosaic virus (TMV) and tomato mosaic virus (ToMV) have been ranked as the most important tomato pathogens (Jones et al. 2016). TMV and ToMV are related viruses belonging to the genus Tobamovirus, family Virgaviridae (Adams et al. 2009). Typically, they have rod-shaped particles of 300 × 18 nm in size, which encapsidate the genomic single-stranded positive-sense RNA. The viral particles are very stable, highly infectious, and can be spread easily via mechanical transmissions through wounds caused by workers or pollinator insects (Okada et al. 2000; Levitzky et al. 2019). Moreover, TMV and ToMV can also be transmitted by tomato seeds, one of the most important sources of infection (Dombrovsky and Smith 2017). All of these viral properties make the control of tobamovirus infections difficult.
Till now, breeding and use of resistant S. lycopersicum cultivars proved to be the most effective strategy to control tobamoviruses. Three tobamovirus resistance genes [Tm-1, Tm-2, and Tm-22 (Tm-2a)] have been transferred into S. lycopersicum via crossing with wild tomato species. The Tm-1 derived from S. habrochaites (PI 126,445) is an incompletely dominant gene that suppresses virus replication (Holmes 1954; Pelham 1972; Fraser et al. 1980). The Tm-2 and Tm-22 alleles were introgressed from S. peruvianum (PI 126,926, PI 128,650), conferring complete dominant resistance based on the hypersensitive reaction of the host plant (Laterrot and Pecaut 1969; Alexander 1963; Schroeder et al. 1967; Pfitzner 2006).
Resistant breaking strains of TMV or ToMV have been detected for decades (Betti et al. 1997; Calder and Palukaitis 1992), but these strains did not spread widely in tomato crops until now. However, a new tobamovirus first isolated in Jordan and named as Tomato brown rugose fruit virus (ToBRFV) (Salem et al. 2016) caused a “pandemic alert” in Europe (EPPO Global Database 2016).
ToBRFV is a high-risk pathogen due to its rapid distribution across several continents (Africa, Asia, and North America) and its ability to overcome the resistance genes Tm-2 and Tm-22 introgressed into tomato during the past decades (Luria et al. 2017). The virus infection can occur by seed transmission as primary inoculum and pollen transmission of bumblebee (Bombus terrestris) (Dombrovsky and Smith 2017; Levitzky et al. 2019; Davino et al. 2020; Salem et al. 2021). Symptoms caused by ToBRFV vary greatly depending on the genotype, age, and environment of the infected tomato. Foliar symptoms usually appear as chlorosis, mosaic patterns, and mottling, occasionally accompanied with leaf narrowing. Fruits of diseased plants show yellow or brown wrinkled (rugose) patches rendering them unmarketable (Salem et al. 2016; Luria et al. 2017). Besides tomato, ToBRFV is able to infect sweet pepper (Capsicum annuum) (Salem et al. 2020; Panno et al. 2020). Because the resistance genes are not active to ToBRFV, there is an urgent demand to find new sources of resistance. The present study aimed to screen for the susceptibility and resistance of a wide range of wild tomatoes and some Solanum relatives to ToBRFV.
Materials and methods
Plant materials
A total of 636 plant accessions belonging to different Solanum (sections Lycopersicon and Juglandifolia) species such as S. arcanum (9); S. cheesmaniae (21); S. chilense (99); S. chmielewskii (10); S. corneliomulleri (26); S. galapagense (11); S. habrochaites (22); S. huaylasense (9); S. juglandifolium (3); S. lycopersicum (81); S. neoricki (16); S. ochranthum (5); S. pennellii (18); S. peruvianum (43); S. pimpinellifolium (256); and S. sitiens (7) were investigated. The following accessions were used as a control: S. habrochaites (PI 126,445; original source of the Tm-1 gene), S. peruvianum (PI 126,926; source of the Tm-2 gene, and PI 128,650; source of the Tm-22 gene), S. lycopersicum (LA1221; carrying the introgressed Tm-22 gene), and the susceptible cultivar S. lycopersicum (‘Ceglédi’; +/+). The seeds of Solanum species were kindly supplied by the Tomato Genetic Resources Centre (University of California, Davis), United States Department of Agriculture Agricultural Research Service (Beltsville, Maryland) and MATE (Hungarian University of Agriculture and Life Sciences). The seeds were sown in fertilized Klasmann Traysubstrate soil, and potted plants were grown in an insect-proofed glasshouse at 24 ± 2 °C temperature, 14/10 h photoperiod, and 50–70% relative humidity.
Virus isolates and plant inoculation
A Jordanian ToBRFV isolate (GenBank acc.no. MZ323110) was employed in the present work. Tobacco mosaic virus (TMV-U1) and tomato mosaic virus (ToMV-DH) isolates were received from the virus collection of MATE (Hungarian University of Agriculture and Life Sciences) kindly provided by Pál Salamon. ToBRFV, TMV, and ToMV were transmitted through a single local lesion in Nicotiana glutinosa and propagated on N. tabacum cv. Samsun. Inocula were prepared by grinding infected tobacco leaves in sterile 0.01 M phosphate buffer pH 7.0 (1:5 w/v). The sap was then filtered through a cheesecloth, and the extract was stored in aliquots at − 20 C0 for inoculation through this work.
For mechanical transmission, virus inoculum was rubbed onto carborundum-dusted lower leaves of young tomato and tobacco test plants. At least 3–10 plants from each accession were inoculated at 3–4 true leaf stage. Local and systemic symptoms were evaluated 3–4 weeks post-inoculation (wpi). In each inoculation experiment, the infectivity of the inocula was assayed using N. glutinosa and/or N. tabacum cv. Xanthi-nc as local lesion test plants. All greenhouse and laboratory experiments were carried out in quarantine conditions.
Evaluation of disease symptoms and handling of resistant plants
The disease caused by ToBRFV was assessed in each inoculated plant 2–3 wpi, and the disease severity index (DSI) listed (Table 1) was calculated by the formula developed by (Camara et al. 2013):
where \(\mathrm{DSI}\) = disease severity index; e = class; Re = number of plants in class (e); N = total number of plants.
Symptomless plants (class 0) confirmed as virus-free by bioassay and RT-PCR were predicted to be resistant. These plants were re-inoculated and tested for the presence of the virus again. Three weeks after the second inoculation, the symptomless plants were decapitated. Two weeks later, two lateral shoots from the decapitated plants were inoculated again and checked for the presence of the virus. Simultaneously, one non-inoculated lateral shoot of each plant was cut off, rooted in MS media, propagated in vitro and transferred to pots. Three propagated individuals were inoculated by TMV, ToMV, and ToBRFV, respectively, and assayed for the presence of tobamovirus by bioassay and RT-PCR.
Detection of viruses
The presence or absence of viruses was demonstrated in leaf samples of inoculated plants using bioassay and reverse-transcription polymerase chain reaction (RT-PCR). Samples were taken at 2–3 wpi from newly developed top leaves. The assayed leaves were rinsed with sodium hydroxide (2%) and then with tap water to avoid virus contamination. Bioassays were carried out by rubbing indicator plants (N. glutinosa and N. tabacum cv. Xanthi nc) with leaf extract prepared from inoculated and top leaves of donor tomato plants, respectively. For RT-PCR, RNA was extracted using Promega SV Total RNA extraction kit following the manufacturer’s instructions. Extracted RNA samples were used as a template for complementary DNA (cDNA) transcription oligonucleotide specific for ToBRFV, ToMV, and TMV specific primers. Primer3 computer software (version 4.0.0) was used to design the PCR primers using the ToBRFV (KT383474), ToMV (MH507165) and TMV (FR878069) reference virus genomes. To amplify the coat protein segment, the following primers were used: for ToBRFV F-5894 (5'- GTTCCAAACACAACAAGCTAGA -3') and R-6250 (5'- AAAGTGCATCCGGTTTACAATG -3'), for ToMV F-5894 (5'- GTTTCAAACACAGCAAGCAAGA -3') and R-6250 (5'- CAGACCAACCCAGACATACTTT -3'), and for TMV F-5809 (5'- CTCCATCTCAGTTCGTGTTCTTG -3') and R-6250 (5'- CAAACCAAACCAGAAGAGCTCT -3'). PCR products were detected by electrophoresis in agarose gel (2%, in 0.5 X TBE buffer).
Results
Evaluation of Solanum accessions for the susceptibility and resistance to ToBRFV
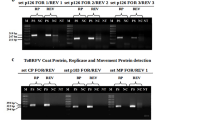
Six hundred thirty-six Solanum accessions were inoculated with ToBRFV and evaluated for symptoms and DSI (Table 1, Supplementary 1). The control accessions, S. peruvianum (PI 126,926; Tm-2, PI 128,650; Tm-22), S. lycopersicum (LA1221; Tm-22), and S. lycopersicum (Ceglédi; +/+), showed DSI ranged between 80 and 100%, whereas S. habrochaites (PI 126,445; Tm-1) plants showed milder symptoms displaying a DSI of 20%. Plants of 603 wild Solanum accessions were susceptible and showed systemic symptoms at different severity levels (Fig. 1, Supplementary 1). Twenty-six accessions of S. pimpinellifolium (LA1301, LA1375, LA1547, LA1579, LA1607, LA1611, LA1612, LA1630, LA1634, LA1661, LA1670, LA1676, LA1679, LA1685, LA1728, LA1924, LA2903, LA2904, LA2982), two accessions of S. habrochaites (LA1559, LA2174), one accession of S. chilense (LA1932), and four accessions of S. lycopersicum var. cerasiforme (LA1456, LA2675, LA2688, LA1385) were found to be tolerant showing no symptoms at all or very mild mosaic symptoms with average disease severity between 0 and 20%. The presence of the virus in tolerant plants was confirmed by using RT-PCR and bioassays (Figs. 2 and 3).
The inoculated and top leaves of S. ochrantum accessions, LA2160, LA2162 and LA2166, remained symptomless following the first, the second and the lateral shoot inoculation by ToBRFV. The presence of the virus has been detected only on inoculated leaves using bioassay. Similar responses were observed on these accessions also inoculated with TMV and ToMV, respectively. The other two S. ochrantum accessions PI 473,498 and PI 230,519 showed distinct responses to ToBRFV, ToMV, and TMV. They were infected only locally by TMV and ToMV but infected locally and systemically by ToBRFV. The systemic reactions of the S. ochrantum accessions PI 473,498 and PI 230,519 were unexpected. They initially expressed mild systemic mosaic symptoms (DSI 20%) at 15 days post-inoculation (dpi) and contained infective virus. However, they recovered from the symptoms (Fig. 4) and the virus could not be detected on their newly emerged symptomless leaves (Table 2, Fig. 5).
Detection of ToBRFV by RT-PCR in resistant S. ochranthum plants. M = molecular marker, 1 = negative control, 2 = LA2160, 3 = LA2162, 4 = LA2166, (5–6) = PI 473,498 and PI 230,519, 7 = positive control samples. Plants 5–6 showed mild mosaic after inoculation, and according to the bioassays on N. tabacum var. Xanthi-nc they contained virus, but later they recovered, and no virus could be detected in their top leaves by bioassays and RT-PCR
Discussion
Several members of the Tobamovirus genus like TMV and ToMV have been known for a long time as harmful pathogens of tomato. These mechanically and seed transmitted stable viruses were successfully controlled using resistant cultivars and hybrids carrying the well-known resistance genes Tm-1, Tm-2, and Tm-22 (Soost 1963; Alexander 1963; Pfitzner 2006). Although some TMV and ToMV mutants have been reported to overcome the resistance conferred by these genes, they did not spread widely and no serious yield losses were reported (Betti et al. 1997; Calder and Palukaitis 1992). However, ToBRFV, a recently emerged plant virus (Salem et al. 2016), has been demonstrated to infect all the known genotypes carrying characterized resistance genes and caused worldwide panic among seed companies and tomato producers (Luria et al. 2017; Dombrovsky and Smith 2017). Resistance to this new virus has been demonstrated in some genotypes of S. pimpinellifolium, S. lycopersicum and S. habrochaites (Hamelink et al. 2019; Ashkenazi et al. 2020; Ykema et al. 2020), while tolerance to ToBRFV has been found in a genotype S. lycopersicum and S. pimpinellifolium (Ashkenazi et al. 2018; Zinger et al. 2021).
Studying the reaction of 636 accessions of 16 species, we found that most of them were susceptible, including the accessions of S. arcanum, S. chmielewskii, S. huaylasense, S. juglandifolium, S. sitiens, and S. ochranthum (Supplementary 1). To the best of our knowledge, the last-mentioned six species have never been studied as hosts or non-host of ToBRFV, so they can be indicated as new experimental hosts of this virus.
Several accessions were found to be tolerant (listed in Supplementary 1). They could be infected by ToBRFV, but did not show any systemic symptoms. A similar tolerant reaction has been identified in S. lycopersicum and S. pimpinellifolium (Ashkenazi et al. 2018; Zinger et al. 2021). However, in addition to the cultivated lines of S. lycopersicum var. cerasiforme and S. pimpinellifolium, we also demonstrated tolerance in accessions of the wild tomato plants of S. habrochaites and S. chilense.
The reaction of S. ochranthum, a close relative to tomato, varied greatly. Two accessions (PI 473,498 and PI 230,519) showed transitional mild systemic mosaic symptoms followed by total recovery on the new apical leaves. While ToBRFV could be detected by bioassays in the mosaic displaying leaves, no virus was present later on the new symptomless top leaves, indicating either the arrest of virus movement or the very strict control of virus replication. Interestingly, similar recovery from disease, including vanishing of symptoms and lack of detectable viruses, has been already reported in S. ochrantum when inoculated with the potexvirus, pepino mosaic virus (Soler-Aleixandre et al. 2007).
In contrast to the accessions PI 473,498 and PI 230,519, plants of three S. ochranthum accessions (LA2160, LA2162, and LA2166) inoculated with ToBRFV remained symptomless both locally and systemically and the virus could be detected only on the inoculated leaves of these plants. These results proved their high level of resistance to ToBRFV. High resistance of S. ochranthum was also found against TMV and ToMV, which suggest the same genetic background of resistance to different tobamoviruses in these plants. Reactions of S. ochranthum have been studied so far to cucumber mosaic virus (CMV) and pepino mosaic virus (PepMV) (Rick 1988; Soler-Aleixandre et al. 2007). This is the first paper dealing with the reactions of S. ochranthum to tobamoviruses. It would be of special interest to know whether S. ochrantum is susceptible or resistant to other important solanaceous pathogenic tobamoviruses like tomato mild mottle virus (ToMMV) or obuda pepper virus (ObPV).
The transfer to cultivated tomato of this high level of resistance of S. ochrantum to ToBRV is difficult due to the sexual incompatibility between S. ochranthum and S. lycopersicum or other closely related tomato species. A potential alternative to surpass this genetic barrier will be the use of somatic hybridization among accessions of these species (Rick 1979; Rick and Chetelat 1995; Pertuzé et al. 2002; Kole 2011).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Adams MJ, Antoniw JF, Kreuze J (2009) Virgaviridae: a new family of rod-shaped plant viruses. Arch Virol 154:1967–1972. https://doi.org/10.1007/s00705-009-0506-6
Alexander LJ (1963) Transfer of a dominant type of resistance to 4 known Ohio pathogenic strains of tobacco mosaic virus (TMV), from Lycopersicon Peruvianum to Lycopersicon esculentum. Phytopathology 53:869
Ashkenazi V, Rotem Y, Ecker R, Nashilevitz S, Barom N (2018) Tolerance in plants of Solanum lycopersicum to the tobamovirus tomato brown rugose fruit virus (TBRFV). Patentscope 38 pp. https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2018219941
Ashkenazi V, Rotem Y, Ecker R, Nashilevitz S, Barom N (2020) Resistance in plants of Solanum lycopersicum to the tobamovirus tomato brown rugose fruit virus. Patentscope 60 pp. https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2020249798&tab=SEARCHREPORT
Betti L, Marini F, Marani F, Cuffiani M, Rabiti AL, Canova A (1997) A TMV strain overcoming both Tm-2 and Tm-22 resistance genes in tomato. Phytopathol Mediterr 36:24–30
Calder VL, Palukaitis P (1992) Nucleotide sequence analysis of the movement genes of resistance breaking strains of tomato mosaic virus movement genes, encoding 30K proteins, of two resistance breaking strains of tomato mosaic virus. J Gen Plant Pathol 73:165–168. https://doi.org/10.1099/0022-1317-73-1-165
Camara M, Mbaye AA, Noba K, Samb PI, Diao S, Cilas C (2013) Field screening of tomato genotypes for resistance to tomato yellow leaf curl virus (TYLCV) disease in Senegal. Crop Prot 44:59–65. https://doi.org/10.1016/j.cropro.2012.10.007
Davino S, Caruso AG, Bertacca S, Barone S, Panno S (2020) Tomato brown rugose fruit virus: seed transmission rate and efficacy of different seed disinfection treatments. Plants 9:1615. https://doi.org/10.3390/plants9111615
Dombrovsky A, Smith E (2017) Seed transmission of tobamoviruses: aspects of global disease distribution. In: Jimenez-Lopez JC (ed) Seed biology. IntechOpen, London, pp 234–260
EPPO Global Database (2016) New data on quarantine pests and pests of the EPPO Alert List. https://gd.eppo.int/taxon/TOBRFV
Fraser RSS, Loughlin SAR, Connor JC (1980) Resistance to tobacco mosaic virus in tomato: effects of the Tm-1 gene on symptom formation and multiplication of virus strain 1. J Gen Virol 50:221–224. https://doi.org/10.1099/0022-1317-50-1-221
Hamelink R, Kalisvaart J, Rashidi H (2019) TBRFV resistant tomato plant. Patentscope 34 pp. https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2019110130&tab=PCTBIBLIO&_cid=P10-KPB4P1-64395-1
Heuvelink E (2018) Tomatoes (crop production science in horticulture 27), 2nd edn. CABI, Leusden
Holmes FO (1954) Inheritance of resistance to infection by tobacco-mosaic virus in tomato. Phytopathology 44:640–642
Jones JB, Zitter TA, Miller SA (2016) Compendium of tomato diseases and pests, 2nd edn. APS Publisher, Saint Paul
Kole C (2011) Wild crop relatives: genomic and breeding resources: vegetables. Springer Science & Business Media, New York
Laterrot H, Pecaut P (1969) Gene Tm-2: new source. Rep Tomato Genet Coop 19:13–14
Levitzky N, Smith E, Lachman O, Luria N, Mizrahi Y, Bakelman H, Sela N, Laskar O, Milrot E, Dombrovsky A (2019) The bumblebee Bombus terrestris carries a primary inoculum of tomato brown rugose fruit virus contributing to disease spread in tomatoes. PLoS ONE 14(1):e0210871. https://doi.org/10.1371/journal.pone.0210871
Luria N, Smith E, Reingold V, Bekelman I, Lapidot M, Levin I, Elad N, Tam Y, Sela N, Abu-Ras A, Ezra N, Haberman A, Yitzhak L, Lachman O, Dombrovsky A (2017) A new Israeli Tobamovirus isolate infects tomato plants harboring Tm-22 resistance genes. PLoS ONE 12:1–19. https://doi.org/10.1371/journal.pone.0170429
Okada K, Kusakari S, Kawaratani M, Negoro J, Satoshi TO, Osaki T (2000) Tobacco mosaic virus is transmissible from tomato to tomato by pollinating bumblebees. J Gen Plant Pathol 66:71–74. https://doi.org/10.1007/PL00012924
Panno S, Caruso AG, Blanco G, Davino S (2020) First report of tomato brown rugose fruit virus infecting sweet pepper in Italy. New Dis Rep 41:588–2044. https://doi.org/10.5197/j.2044-0588.2020.041.020
Pelham J (1972) Strain-genotype interaction of tobacco mosaic virus in tomato. Ann Appl Biol 71:219–228
Pertuzé RA, Ji Y, Chetelat RT (2002) Comparative linkage map of the Solanum lycopersicoides and S. sitiens genomes and their differentiation from tomato. Genome 45:1003–1012. https://doi.org/10.1139/g02-066
Pfitzner AJP (2006) Resistance to tobacco mosaic virus and tomato mosaic virus in tomato. In: Loebenstein G, Carr JP (eds) Natural resistance mechanisms of plants to viruses. Springer, Dordrecht
Rick CM (1979) Biosystematic studies in Lycopersicon and closely related species of Solanum. In: Linnean Society symposium series pp 667–679
Rick CM (1988) Tomato-like nightshades: affinities, autoecology, and breeders’ opportunities. Econ Bot 42:145–154
Rick CM, Chetelat RT (1995) Utilization of related wild species for tomato improvement. Acta Hortic 412:21–38. https://doi.org/10.17660/ActaHortic.1995.412.1
Salem N, Mansour A, Ciuffo M, Falk BW, Turina M (2016) A new tobamovirus infecting tomato crops in Jordan. Arch Virol 161:503–506. https://doi.org/10.1007/s00705-015-2677-7
Salem NM, Cao MJ, Odeh S, Turina M, Tahzima R (2020) First report of tobacco mild green mosaic virus and tomato brown rugose fruit virus infecting Capsicum annuum in Jordan. Plant Dis 104:601
Salem NM, Sulaiman A, Samarah N, Turina M, Vallino M (2021) Localization and mechanical transmission of tomato brown rugose fruit virus in tomato seeds. Plant Dis. https://doi.org/10.1094/PDIS-11-20-2413-RE
Schroeder WT, Provvidenti R, Robinson RW (1967) Incubation temperature and virus strains important in evaluating tomato genotypes for tobacco mosaic virus reactions. Tomato Genet Coop Rep 17:47–48
Soler-Aleixandre S, López C, Cebolla-Cornejo J, Nuez F (2007) Sources of resistance to pepino mosaic virus (PepMV) in tomato. HortScience 42:40–45. https://doi.org/10.1094/PDIS-91-6-0749
Soost RK (1963) Hybrid tomato resistant to tobacco mosaic virus. J Hered 54:241–244. https://doi.org/10.1093/oxfordjournals.jhered.a107258
Ykema M, Verweij CW, De La Fuente Van Bentem S (2020) Tomato plant resistant to tomato brown rugose fruit virus. Patentscope 143 pp. https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2020147921&tab=PCTBIBLIO
Zinger A, Lapidot M, Harel A, Doron-Faigenboim A, Gelbart D, Levin I (2021) Identification and mapping of tomato genome loci controlling tolerance and resistance to tomato brown rugose fruit virus. Plants 10:179. https://doi.org/10.3390/plants10010179
Acknowledgements
The authors would like to thank Lilla Hajnik for her help in molecular work and Peter Kalo for editing the manuscript
Funding
Open access funding provided by Hungarian University of Agriculture and Life Sciences. This study was supported by the Stipendium Hungaricum scholarship program supervised by the Ministry of Foreign Affairs and Trade of Hungary and managed by the Tempus Public Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
The authors have consented to the publication of the current version of the article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jewehan, A., Salem, N., Tóth, Z. et al. Screening of Solanum (sections Lycopersicon and Juglandifolia) germplasm for reactions to the tomato brown rugose fruit virus (ToBRFV). J Plant Dis Prot 129, 117–123 (2022). https://doi.org/10.1007/s41348-021-00535-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-021-00535-x