Abstract
Mediterranean fish species living along Italian (Gaeta) and Egyptian (Alexandria) coasts were analyzed using DNA barcodes for molecular identification. Mitochondrial Cytochrome c Oxidase subunit 1 (COI) gene was sequenced from 31 different marine species to test whether the morphology-based assignment of individuals into 19 families, 6 orders was supported by DNA-based species delimitation and Neighbour Joining cladogram. All COI rRNA gene barcodes were matched with reference sequences of expected species, according to morphological identification. Neighbour joining tree was drawn based on COI rRNA gene and the majority of specimens clustered in agreement with their taxonomic classification. Our results updated Mediterranean edible fish knowledge providing graphical resources, taxonomical and bioinformatics references, improving the genetic fish database and the basic molecular information to strengthen the science–policy interface for biodiversity and ecosystem services as conservation, blue economy, and long-term human well-being.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Mediterranean Sea has a significant economic importance for its commercial fish trading, requiring periodic checks on the diversity of marine communities and quality of its marketable fish products. The catalog of species is an ancient idea, which began in 1770 by Linnaeus, basing on morphological characters (for review see Charmantier and Müller-Wille 2014). To help non-experts in the morphological identification process, a universal marker called ‘‘DNA barcode’’ has been used in the last decade (Waugh 2007). The DNA barcode is the sequence of the ‘‘Folmer fragment’’ (Folmer et al. 1994), a polymorphic part of the mitochondrial cytochrome oxidase subunit I gene (COI), which can be used to identify closely related species as well as higher taxa in many animal phyla (Hebert et al. 2003a). It can help to discern between similar species (Hebert et al. 2003b) allowing an accurate analysis of biodiversity differentiating between doubtful species, subspecies, hybrids and species still unknown (Mytilineou et al. 2016). Several studies highlight the importance to monitor ecological/environmental problems: the technique is a useful tool in the ecological and environmental monitoring to define what are the areal distribution of these species and their evolution or control which species are at risk of extinction and to suggest possible approaches (Scheffers et al. 2012). Furthermore, through the technique of barcoding it is possible to identify a species from a small portion or living and/or ancient tissue (Puillandre et al. 2012), undesired materials inside of foodstuffs or commercial products and it is also possible to recognize the species at all stages of life, such as eggs and larvae (Briski et al. 2011). Since European food law aims to ensure a high level of traceability and food security (Reg EC 178/2002; Reg EC 1420/13) and in Egypt, as in Europe, a clear label must be added to all fish products, including the taxonomic names for species and family (directive ES:3494/2005; subject 3/7/3), barcoding can be very useful to provide label species-specific information. In this sense, barcoding reveals commercial frauds, particularly in seafood fishing industry, where species more expensive can be exchanged with species of lesser economic and nutritional value, sometimes causing the occurrence of allergies and poisonings (FAO 2003; Trocchia et al. 2015). Barcoding monitors preserve biodiversity not only for the animal chain conservation but also for the species sustainability since biodiversity is a network of elements, connections, and functions characterizing the life, where the man is an integral part of it and he depends closely on it (Patel 2014). For this reason, biodiversity must be known and regularly cataloged (Crocetta et al. 2015). It requires a heavy commitment because the biological diversity of an environment is characterized by a considerable biological wealth, hosting almost all the animal phyla, including those who do not live on land or in fresh water, welcoming about 10 million species, substantially an higher amount of terrestrial environments (Bianchi and Morri 2000; Appeltans et al. 2011; Bilecenoglu et al. 2013). The environment is constantly changing as well as ecosystems due to phenomena of extinction, hybridization and non-native species resettlement (Occhipinti-Ambrogi et al. 2011; Guerriero et al. 2010; Hui 2013). That inexorable change of the articulated trophic networks which regulate biodiversity and the blue economy (Guerriero 2013) makes the species-cataloging process considerably more complex. Through the technique of barcoding it is possible to identify a species and detect the corresponding species-specific content of antioxidants. Between foods of animal origin, fishes are the ones which contain a higher content of alpha tocopherol (vitamin E) but difference can be in the amount present in relation to the fish species (Kris-Etherton et al. 2002; Guerriero et al. 2008). Among the vitamins, alpha tocopherol is the most studied because it is involved in many biological functions and it has an important role as antioxidant factor, preventing the oxidation of polyunsaturated fatty acids (Esterbauer et al. 1991; Guerriero et al. 2002, 2004). In addition to its activities as an antioxidant, α-tocopherol is involved in immunity process and in the transmission of signals between cells in the regulation of gene expression and other metabolic processes (Rimbach et al. 2002) resulting in several benefits to human health: it is a powerful immune stimulant; acts in the prevention and treatment of heart disease, favoring the dissolution of any blood clots in the arteries and it is able to decrease the rate of cholesterol in the bloodstream, thus avoiding deposits of fat; it relieves pain due to varicose veins, headaches, and nervous system problems; it appears to be implicated in the prevention of cancer due to its antioxidant properties (Halver 2002). Recently, barcoding is used for biodiversity evolution studies too (Lahaye et al. 2008). This technique permits, in fact, a quick enumeration of organisms present in a given habitat and detects any variation phenomena of biodiversity in time. The evaluation of the increase or decrease of biodiversity helps to estimate, indirectly, the health of ecosystems (Lahaye et al. 2008). Aim of our study was to discriminate the Mediterranean edible fish along Italian and Egyptian coast, obtained in the framework of the memorandum of understanding between Federico II University, Napoli (Italy) and the National Research Centre of Excellence for Advanced Science (CEAS) in Cairo (Egypt) to improve the knowledge of Mediterranean marine communities’ diversity, their genetic fish database, and to analyze the sequences of rRNA gene fragments obtained.
Materials and methods
Sampling
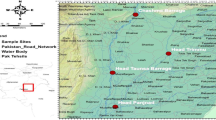
Samples of Gaeta Gulf and Alexandria Harbor along Mediterranean Sea (Fig. 1) have been kindly provided by Italian and Egyptian local fisherman and their pictures (Fig. 2A–C) by the scuba diver Dr. Adrıano Madonna from March 2013 to May 2015. Each fish species was examined with three specimens/species regardless of the samples received. Morphological identification of the specimens was performed according to fish identification keys of the World Register of Marine Species, WoRMS (https://readtiger.com/wkp/en/World_Register_of_Marine_Species#Contents) and the Integrated Taxonomic Information System (http://www.itis.gov). After morphological examination, muscle tissue samples were dissected from each specimen and stored at −20°C until molecular processing.
A Panel representing analyzed species, photographed in nature: a Anguilla anguilla; b Conger conger; c Aulopus filamentosus; d Sardina pilchardus; e Engraulis encrasicolus; f Seriola dumerili; g Trachurus mediterraneus; h Centracanthus cirrus; i Xyrichtys novacula; j Dicentrarchus labrax. B Panel representing analyzed species, photographed in nature: a Dicentrarchus punctatus; b Mullus barbatus; c Mullus surmuletus; d Sciaena umbra; e Scomber scombrus; f Thunnus alalunga; g Serranus cabrilla; h Boops boops; i Dentex dentex; j Diplodus sargus; k Lithognathus mormyrus. C Panel representing analyzed species, photographed in nature: a Pagellus erythrinus; b Sparus aurata; c Sphyraena sphyraena; d Sphyraena viridensis; e Echiichthys vipera; f Xiphias gladius; g Lepidorhombus boscii; h Solea solea; i Chelidonichthys lucerna; j Trigla lyra
DNA isolation
DNA isolation was performed in muscle of three specimens from each species of fish (aliquots of 100 mg) as reported in Di Finizio et al. (2007), with a phenol/chloroform standard method using autoclaved glassware and equipment. About 100 mg of ground freeze-dried tissues were mixed in a DNA extraction buffer (50 mM NaCl, 10 mM EDTA, and 10 mM Tris base) and the cells were lysed by adding 2% sodium dodecyl sulfate. The RNA was removed adding RNAse (10 mg/mL) followed by incubation at 37 °C for 30 min. Proteinase K was added (0,5 mg/mL) to remove protein and the samples were incubated for 1 h at 37 °C in a shaking water bath. The extracts were further purified extracting twice with phenol:chloroform:isoamyl alcohol (25:24:1 v/v) and centrifuging at 10,000×g for 15 min at 4 °C. The upper aqueous layer was transferred into a new micro-centrifuge tube and the DNA was precipitate by adding 1/10th volume of 3 M sodium acetate at pH 5.2 and two volumes of 100% chilled ethanol to each sample and mixed centrifuged at 15,000×g for 30 min at 4 °C. The pellet was washed with 70% ethanol, air dried, and finally re-suspended in 50 μl of sterilized deionized water. Optical density (OD) of each sample was measured at 260 and 280 nm, respectively, by UV spectrophotometer (Biochrom Libra S12), and the purity of DNA was measured by the OD260/OD280 ratio (ideal ratio = 1.7–2.0), and the quality was observed by electrophoresis on a 0.8% agarose gel and visualized under UV light (Di Finizio et al. 2007).
PCR amplification and sequencing
DNA barcoding region was amplified, from the 5′ region of the mitochondrial cytochrome c oxidase subunit I (COI) gene, as previously published (Di Finizio et al. 2007) using the following primers: COI_UP (5′-ACTTCAGGGTGACCGAAGAATCAGAA-3′) and COI_DW (5′-ATCTTTGGTGCATGAGCAGGAATAGT-3′) (Ward et al. 2005); 12S, 16S, and cytb, rRNA gene fragments (cytb_UP (5′- CCATCCAACATCTCAGCATGATGAAA-3) cytb_DW (5′- GCCCCTCAGAATGATATTTGTCCTCA -3′) (Kocher et al. 1989; Madonna et al. 2015), 12S_UP (5′-AAACTGGGATTAGATACCCCACTATc), 16S_UP (5′-GCCTGTTTATCAAAAACAT), 16S_DW (5′-CCGGTCTGAACTCAGATCACGT-3′), 12S_DW (5′-GAGGGTGACGGGCGGTGTGT-3′reported in Di Finizio et al. 2007; Guerriero et al. 2010) and PCRs were performed in a Techgene Thermal Cycler (Thecne Ltd., Cambridge, UK). The amplifications were carried out in a reaction buffer containing 50 mM KCl, 10 mM Tris/HCl, pH 9.0; 10 mM NaCl; 0.01 mM EDTA; 2.5 mM of each dNTP; 1 μM of each primer; 10 ng of template DNA; 0.5 unit of Taq DNA polymerase (Invitrogen, Milan, Italy). Conditions of PCR amplification were as follows: COI rRNA gene (35 cycles, with a denaturation at 94 °C for 50 s, annealing at 54 °C for 50 s, and extension at 72 °C for 1 min); 12S and 16S rRNA gene (initial denaturation step at 95 °C for 10 min, followed by 35 amplification cycles, with denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min and 30 s, followed by a final extension step at 72 °C for 7 min); cytb rRNA gene (35 cycles, with denaturation at 93 °C for 1 min, annealing at 50 °C for 1 min, and extension at 72 °C for 2 min). At the end of the incubation, 5 μl of PCR products were separated by electrophoresis through 2% agarose gel and visualized under UV light. A 100-bp ladder (Invitrogen, Milan, Italy) was used to estimate the fragment size of the amplicons generated. Amplified DNA was desalted and purified with Microcon 100 spin columns (Millipore-Amicon, Belford, MA, USA) according to the manufacturer’s instructions, and sequenced using Big Dye TM Terminator Cycle Sequencing Chemistry (Applied Biosystems, Foster City, CA, USA) in an automatic capillarity sequencer (ABI 310 Genetic Analizer; Applied Biosystems).
Sequence alignment and analysis
Nucleotide sequences were aligned using Chromas 1.45 vs (Technelysium 186 Pty, Tewantin, Australia) and BioEdit (Tom Hall Ibis Therapeutics, Rutherford Road Carlsbad, CA, USA) software and compared with GenBank sequences using FASTA (Mount 2007) to obtain the best match found in GenBank database to confirm species identification and quantify a possible divergence/similarity from query sequences.
Multiple alignments were performed with the program Clustal Omega (Sievers and Higgins 2013) to ensure that all sequences of COI marker gene provide a homologous fragment and to detect the percentage of similarity found between COI rRNA gene sequences underlining a possible genetic divergence between them (Ward 2012). COI rRNA gene sequences were translated into amino acids with the online program ExPASy translation tool (http://web.expasy.org/translate) to exclude sequencing errors and to avoid the inclusion of pseudogene sequences in the datasets.
Cladograms reporting genetic distance were realized with MEGA6 software (Tamura et al. 2013), using Neighbour Joining and 1000 bootstrap replicates.
Percentage of barcoded species were obtained by dividing the number of species/order having the accession number in the database (GenBank) and the total number of species collected for the same order, multiplied by 100.
Results
Barcoding sequences and alignment analysis
DNA isolation from muscle of fish constitutes a more efficient template, indicating a good yield of PCR products. The described set of different primers successfully has amplified the corresponding mitochondrial region fragments examined (data not shown). The PCR products isolated from gel were sequenced. All sequences, subjected to FASTA searches against the NCBI DNA database, were classified according to the sequences they aligned to with the highest identity confirming the morphological identification of all 93 specimens representing 31 species, 28 genera, 19 families, and 6 orders (Table 1). The length (base pair) of sequences obtained for each analyzed species and the percentage of similarity between sequence/species are reported in Table 2. Genetic distances within species in terms of number of base substitution evidenced by the COI rRNA gene divergence are 2% as demonstrated by the similarity percentage of all specimen sequences around. The percentage of COI sequence variability between Italian and Egyptian specimen sequences estimated is very low and between 1 and 1.5. Trachurus mediterraneus Italian and Egyptian sequences have 98.5% of COI variability, Dicentrarchus punctatus 99%, Solea solea, Dentex dentex and Diplodus sargus have 99% for Italian sequences and 98.5 for Egyptian ones (Table 2).
To give a complete vision of the processed data so far, a phylogenetic analysis was conducted to classify the sequences and to graphically confirm the homology relationship. The NJ tree including 31 species is given in Fig. 3 with indication of distance scale. The species of each order examined are grouped into different clades with low distance. Most of the species belong to Perciformes. Furthermore, each species captured is closely related, with the exception of Sphyraena sphyraena, Sphyraena viridensis, Anguilla anguilla, Conger conger, which are species with deviations at higher levels (order). Echiichthys vipera is the species with the lowest family ties among all represented species. Cladogram demonstrates the presence of species of the same genus among the listed species, such as Mullus barbatus and Mullus surmuletus, Dicentrarchus punctatus and Dicentrarchus labrax, Dentex dentex and Pagellus erythrinus. Highest genetic distance within species was calculated for Echiichthys vipera (0.11) and the lowest genetic distance (except intraspecific distances) was between Spyraena sphyraena and Spyraena viridensis (0.06). Detailed information on mean genetic distances within species is shown in Fig. 3. All the specimens of the same species were clustered together (data not shown).
Neighbour Joining (NJ) trees, obtained by alignment of COI sequences for each considered species. Bootstrap support, calculated from 1000 replicates, is shown at each node. Yellow Anguilliformes, orange Aulopioformes, brown Clupeiformes, blue Perciformes, green Pleuronectiformes, grey Scorpaeniformes
Percentage of barcoded species
Table 3 shows the potentiality to improve the database for species collected by the difference in the percentage of barcoding detected for each order. In particular, on the total of the examined species in the present work, the 5% of species belonging to Perciformes sequences appears to be missing for the cytb rRNA gene fragment, while as for the 16S rRNA gene fragment, for the order Anguilliformes, about 50% of the sequences have yet to be published, unlike for the order Perciformes only 5% of the contributions is not present. Finally, for the 12S rRNA gene fragment, the 50% of the sequences are not currently published for species belonging to Anguilliformes, 14% for species belonging to the order Perciformes and 50% for species belonging to the order Scorpaeniformes. The same summary (Table 3) indicates the GenBank accession number of the three mitochondrial gene fragments isolated and, for the first time, sequenced in the following examined species. 12S rRNA for ten species: Aulopus filamentosus (398 bp), Boops boops (391 bp), Diplodus sargus (391 bp), Echiichthys vipera (390 bp), Pagellus erythrinus (395 bp), Sciaena umbra (388 bp), Seriola dumerili (341 bp), Sphyraena viridensis (390 bp), Xiphias gladius (302 bp), and Xyrichtys novacula (390 bp); 16S rRNA for five species: Aulopus filamentosus (507 bp), Engraulis encrasicolus (620 bp), Sciaena umbra (557 bp), Sphyraena viridensis (607 bp), and Xyrichtys novacula (552 bp); and cytb rRNA for two species: Conger conger (300 bp) and Aulopus filamentosus (286 bp) represent, respectively, the new entry.
Discussion
The molecular recognition, offered by barcoding approach, defines the biodiversity of the fish species collected along two different Mediterranean coasts (Fig. 1), the Gaeta Gulf (Italy) and Alexandria Harbor (Egypt). A total of 31 species collected were morphological analyzed, thus a corresponding set of COI rRNA gene (630–663 bp) sequences was obtained and it confirms the sequences reported in Table 1, available at the GenBank data base (http://www.ncbi.nlm.nih.gov/genbank/). The fish variety, their taxonomy, and genetic distances are shown in Figs. 2A–C, 3b and Table 1, respectively. Our visual census permitted to offer helpful graphical resources. The fish species confirm previous and numerous morphological Mediterranean inventory studies in the literature, valuable information with regard to intra-Mediterranean biodiversity and population structure at the family level and also our molecular studies in Gaeta Gulf (Guerriero et al. 2010; Madonna et al. 2015). Our few collections in Alexandria Harbor have confirmed some species already reported in Akel and Philips (2014). In Gaeta Gulf, 71% of the species examined belong to the order Perciformes that includes, as known, approximately 40% of all existing fish species, whereas the Sparidae family is the numerically representative family collected through the research. The universal primer pair used was able to amplify the COI rRNA gene target region without any deletions or insertions. In addition, no stop codons were found after translation of the nucleotide sequences. The applicability of these primers to 31 marine fish species is a significant indicator that DNA barcoding could be used as a global standard for identifying all fish species as demonstrated by the concordance between sequence and morphology. By FASTA analysis of COI rRNA sequences, we detected that the maximum COI query demonstrates the molecular identification efficiency. In general, as shown in Table 2, for the examined species the COI rRNA gene similarity percentage of all specimen sequences is around 98%. Sequences of this unpublished study belong to other fragments of rRNA analyzed after detection of barcoded percentage of the fish examined. Fragments belonging to mitochondrial 12S rRNA gene (n = 10), 16S rRNA gene (n = 5), and cytb rRNA gene (n = 2) were isolated, sequenced and now available at GenBank (Table 3). The sequences provided for Aulopus filamentosus, Sciaena umbra and Conger conger, not previously available in database, enriched fish database permitting eggs, larvae and fish fragments identification as well as the monitoring and surveillance of their substitute.
Genetic relationship between species is shown in NJ tree (Fig. 3). Each species was associated with a specific DNA barcode cluster and the relationship among these species was clearly revealed. Closer species, in terms of genetic divergence, were clustered at the same nodes, for example, Mullus barbatus and Mullus surmuletus, or Dicentrarchus labrax and Dicentrarchus punctatus. The data used for the construction of a phylogenetic tree reported in Fig. 3 estimate the fish examined that possessed distinct COI sequences.
When we examine the NJ tree, there is a clustering pattern that could be informative about phylogenetic relationships (see species of Sparidae, Anguillidae, Triglidae, Sphyraenidae, etc., examined) but is not suitable for a deep phylogenetic resolution, as reported by Ward et al. (2005), since it follows only in part the 655-bp fragment’s length. However, the NJ tree constructed in this study provides a well-matched classification of the current morphological taxonomy, with only minor discrepancies.
Minor deviations were detected on the NJ tree when comparing the order of the following species. Sardina pilchardus and Engraulis encrasicolus are Clupeiformes; Aulopus filamentosus belongs to the Aulopiformes; Lepidorhombus boshii and Solea solea are Pleuronectiformes; Chelidonichthys lucerna and Trigla lyra are Scorpaeniphormes but they clustered with specimens of Perciformes such as Mullus barbatus, Sciaena umbra, Trachurus mediterraneus, Serranus cabrilla, Xyrichtys novacula, Thunnus alalunga, Echiichthys vipera, etc. We excluded immediately the potential contamination or mislabeling because the sequence alignment with COI rRNA gene fragment sequences of species already reported in GenBank resulted in a very high query (over 90%). The clustering pattern is the result of the alignment of sequences and shows that sometimes there is a strict relationship in morphology between species from the same genus such as for Mullus barbatus and Mullus surmuletus, see Fig. 2B, b and c. These two species, belonging to the same genus, are very similar from the morphological point of view: body elongated and slightly compressed laterally; arched dorsal profile, contrary to ventral side which is almost flat; the head is large, tall and short, with a very steep front. The eyes, round and relatively large, are placed at the top, so as to protrude from the dorsal profile of the head. The mouth is terminal, horizontally cut and little protractile, with slightly prominent upper jaw. In the lower part of the mandibular symphysis, there are two tactile barbs. The first dorsal fin has a triangular shape and is composed of seven–eight spiny rays, of which the first is the smallest. The second dorsal fin is inserted about halfway through the body and is formed by one short thorny radius and eight–nine soft rays which decrease rapidly in height. The anal fin is opposite to the second ridge and follows the shape, but it is lower and shorter; it is composed of two spiny rays and six–seven soft ones. The caudal fin (18 rays) is large and forked with equal lobes. The pectoral fins (15–17 rays, longer at the top) are large and developed more than the ventral, which have one thorny radius and five soft. The coloring of the plumage differs in the two species: in Mullus barbatus it tends to be pink, while in Mullus surmuletus it is red, tending to be scarlet, and there is a series of three–four longitudinal golden-yellow rows.
But, there is a close NJ tree relationship also between Xyricththys novacula and Dicentrarchus punctatus (Fig. 2A, i and B, a), which present very different characteristics. In fact, Xyricththys novacula has an oblong body, very compressed on the sides and very high in the front, colored in red or violet greenish, with large scales and ornate with a vertical indent blue. Wavy violet lines are present on the anal fin and tail straight on. The cape is very high with a nearly vertical front profile. The jaws have a single set of teeth, two of which are long and have a sturdy front. The lateral line is interrupted under the last rays of the dorsal fin, the latter is very long and consists of 8–10 rays, the first two of which are somewhat flexible. Dicenthrarchus punctatus, in contrast, has an oblong body and moderately compressed along the sides, colored in gray lead on the back and silvery white on the sides and in the ventral part. The preopercle is notched with the lower edge, provided with plugs placed forward. The ploughshare is completely toothed. The opercle has two plugs placed backwards. The well-evident lateral line is along the sides. For both, the caudal fin is homocercal and unilobed.
At the end, for species included in the NJ tree within Perciformes, but belonging to a different order, such as Lepidorombhus boscii, Solea solea, (Pleuronectiformes; Fig. 2C, g, h), and Chelidonichthys lucerna, Trigla lyra (Scorpaeniformes; Fig. 2C, i, j), we have similar characteristics within the same order, but different if compared to Perciformes. In fact, the two Pleuronectiformes are flatfish with an oblong body and eyes on the right side. The eyes are small and the back is more advanced. The mouth is small and arched; the jaws of the ocular side are devoid of teeth. The dorsal fin begins at the midway between snout and eye until the first ray of the caudal fin. These species are benthic and live in close contact with the sea bed. In contrast, the two Scorpaeniformes present a body that tapers in the second half of the body. Along the lateral line there are several projecting tubules. The head is massive and wide, with slightly concave profile and numerous thorns. The eye is circular and of modest size. The operculum is provided with several pointed appendages. The mouth, wider and lower, shows jaws bands cardiformi teeth. The pectorals are large and fan-shaped. Also in this case, they are benthic species and very different in form, but have some similar characteristics of Perciformes, such as fusiform body, more or less laterally compressed; the scales are ctenoid, rarely cycloid, in some cases absent; fins are usually equipped with some spines rays; simple or multiple dorsal fin with spines rays in the front; ventral fins in thoracic position, formed from no more than six rays, including the first spiniform; caudal fin with a maximum of 17 principal rays; pectoral fins placed at the hips; skull with temporal bones disjointed and sometimes absent; presence of mesethmoid; absence of orbitosphenoid bone; separated from the sub-orbital preopercle; maxillary that does not delimit the oral margin; normally 4–7 branchiostegal rays; lack of intramuscular bones; swim bladder without pneumatic duct. In general, they are pelagic or coastal species.
The only explanation for the clustering pattern obtained can be a strict genetic distance between specimens. We calculated a mean genetic distance of 0.07 specimens from Sparidae, the most representative family of the Perciformes represented in our cladogram. The same thing is made for the other families of Perciformes, such as Mullidae (0.05), Moronidae (0.06), Scombridae (0.08) and Sphyraenidae (0.09). Species within the Perciformes showed a genetic distance range of 0.05–0.13. Ward (2009) indicated that at higher genetic divergence (i.e., 2%), the probability of conspecificity is very low for fish species, whereas in the 2–4% range, congeneric comparisons predominate. If the divergence rate was considered to be more than 5%, the probability of conspecificity becomes <1% and confamilial comparisons become more probable than the congeneric comparisons as the divergence increases.
In addition, the NJ tree was supposed to cluster species. A better resolution of phylogenetic relationship at different taxonomic levels was not the primary target of this COI barcoding study. Work in progress will define better this evolutionary aspect. The cladogram, any case, allowed to discern between species that are phylogenetically related, similar in morphology and edibility degree but less expensive for the economic management, i.e., Dentex dentex (Fig. 2B, i) and Pagellus erythrinus (Fig. 2C, a), two Sparidae that are morphologically and phylogenetically similar, but economically Dentex dentex is more valuable than Pagellus erythrinus.
A better knowledge of the Mediterranean edible fish species identification supported by this approach could be used in enhancing monitoring, conservation, and management. The utilization of molecular markers (such as COI rRNA gene) to identify cases of thermophilic or alien invasions and species at risk of extinction is a promising method that may resolve the status of some issues recorded in the literature. The study of biodiversity is also important because it provides benefits for humans by the regulation of anthropic and natural processes to the production of essential elements, until its contribution to the blue economy, health and general sense of well-being for humans (Díaz et al. 2006; Guerriero et al. 2010; Trocchia et al. 2015). The products of nature are the basis for many industrial activities, so it is evident that the importance of biodiversity takes for humans alin the alimentary industry, as a source of food (Hiddink et al. 2008; Di Finizio et al. 2007). Furthermore, our biodiversity studies help tourism sector too, because a natural environment rich in biodiversity, as well as taking environmental importance, become a real tourist attraction, providing cultural and esthetic benefits (see Madonna et al. 2015). A practical use of barcoding approach updates the database for new species, their discrimination for correct labeling (Di Pinto et al. 2015), to avoid substitution of specimens with less economic value, poisoning, and the onset of allergies (Trocchia et al. 2015) or just to select properly the species with the highest quantity of antioxidants (Guerriero et al. 2008). This species update can contribute to the necessity of periodic checks on the communities’ diversity and catalog (Lahaye et al. 2008; Crocetta et al. 2015) and in particular may become a potential management tool for local administrations to protect Gaeta Gulf and Alexandria Harbor habitats (Scheffers et al. 2012; Madonna et al. 2015) and the health of ecosystems (Lahaye et al. 2008; http://www.rac-spa.org/sites/default/files/doc_cop/biodiversity.pdf).
Conclusion
This work discriminated the Mediterranean edible fish along Italian and Egyptian coast, obtained in the framework of the memorandum of understanding between Federico II University, Napoli (Italy) and the National Research Centre of Excellence for Advanced Science (CEAS) in Cairo (Egypt). First of all, it allowed to categorize by photos for fast morphological recognition and by summary table with taxonomical and bioinformatics references, species sighted in the Gulf of Gaeta and Alexandria Harbor, in the Mediterranean Sea. Furthermore, it granted the opportunity to perform an update of the fish genetic database in GenBank for Mediterranean species useful for their conservation, sustainability, and the blue economy.
References
Akel EHKh, Philips AE (2014) Fisheries and biodiversity of the beach seine catch from the Eastern Harbor, Alexandria, Egypt. Egypt J Aquat Res 40:79–91. doi:10.1016/J.ejar.2014.03.001
Appeltans W, Bouchet P, Boxshall GA, Fauchald K, Gordon DP (2011) World register of marine species. Proceedings of the future of the 21st century ocean: Marine Sciences and European Research Infrastructures, Brest, France, 2012, vol 28, p 30. http://www.marinespecies.org/. Accessed 12 Jan 2017
Bianchi CN, Morri C (2000) Marine biodiversity of the Mediterranean Sea: situation, problems and prospects for future research. Mar Pollut Bull 40:367–376. doi:10.1016/S0025-326X(00)00027-8
Bilecenoglu M, Alfaya JEF, Azzurro E, Baldacconi R, Boyaci YÖ, Circosta V, Compagno LJV, Coppola F, Deidun A, Durgham H, Durucan F, Erguden D, Fernandez-Alvarez FÁ, Gianguzza P (2013) New Mediterranean Marine biodiversity records. Medit Mar Sci 14:463–480. doi:10.12681/mms.676
Briski E, Cristescu ME, Bailey SA, MacIsaac HJ (2011) Use of DNA barcoding to detect invertebrate invasive species from diapausing eggs. Biol Invasions 13:1325–1340. doi:10.1007/s10530-010-9892-7
Charmantier I, Müller-Wille S (2014) Carl Linnaeus’s botanical paper slips (1767–1773). Intellect His Rev 24:215–238. doi:10.1080/17496977.2014.914643
Crocetta F, Agius D, Balistreri P, Bariche M, Bayhan YK, Çakir M, Ciriaco S, Corsini-Foka M, Deidun A, El Zrelli R, Ergüden D, Evans J, Ghelia M, Giavasi M, Kleitou P, Kondylatos G, Lipej L, Mifsud C, Özvarol Y, Pagano A, Portelli P, Poursanidis D, Rabaoui L, Schembri PJ, Taşkin E, Tiralongo F, Zenetos A (2015) New Mediterranean biodiversity records. Medit Mar Sci 16:682–702. doi:10.12681/mms.1477
Di Finizio A, Guerriero G, Russo GL, Ciarcia G (2007) Identification of gadoid species (Pisces, Gadidae) by sequencing and PCR-RFLP analysis of mitochondrial 12S and 16S rRNA gene fragments. Eur Food Res Technol 225:337–344. doi:10.1007/s00217-006-0420-z
Di Pinto A, Marchetti P, Mottola A, Bozzo G, Bonerba E, Ceci E, Bottaro M, Tantillo G (2015) Species identification in fish fillet products using DNA barcoding. Fish Res 170:9–13. doi:10.1016/j.fishres.2015.05.006
Díaz S, Fargione J, Chapin FS, Tilman D (2006) Biodiversity loss threatens human well-being. PLoS Biol 4(8):e277. doi:10.1371/journal.pbio.0040277
Esterbauer H, Dieber-Rotheneder M, Striegl G, Waeg G (1991) Role of vitamin E in preventing the oxidation of low-density lipoprotein. Am J Clin Nutr 53:314S–321S. http://ajcn.nutrition.org/content/53/1/314S
FAO (2003) Report of the expert conservation and international fish trade and food security. ftp://ftp.fao.org/docrep/fao/006/y4961e/y4961E00.pdf. Accessed 12 Jan 2017
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3:294–299
Guerriero G (2013) Non-native species and biodiversity assessment. J Biodivers Endanger Species 1:e104. doi:10.4172/jbes.1000e104
Guerriero G, Di Finizio A, Ciarcia G (2002) Stress-induced changes of plasma antioxidants of aquacultured sea bass, Dicentrarchus labrax. J Comp Biochem Physiol A 132:205–211. doi:10.1016/S1095-6433(01)00549-9
Guerriero G, Ferro R, Russo GL, Ciarcia G (2004) Vitamin E in early stages of sea bass (Dicentrarchus labrax) development. Comp Biochem Physiol A 138:435–439. doi:10.1016/j.cbpb.2004.06.003
Guerriero G, Cuollo M, Agnisola C, Rampacci M, Ciarcia G (2008) α Tocopherol antioxidative resource variations in fish muscle tissue. J Clin Biochem Nutr 43:440–444. http://sfrrj.umin.jp/JCBN.htm
Guerriero G, Di Finizio A, Ciarcia G (2010) Biological pollution: molecular identification of non-native species in the Central Tyrrhenian Sea. Catrina 5:41–47. http://eses-catrina.com/files/06.pdf
Halver JE (2002) The vitamins. In: Halver JE, Hardy RW (eds) Fish nutrition. Academic, San Diego, pp 61–141. doi:10.1016/B978-012319652-1/50003-3
Hebert PDN, Ratnasingham S, de Waard JR (2003a) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Roy Soc Biol Sci 270:S96–S99. doi:10.1098/rsbl.2003.0025
Hebert PDN, Cywinska A, Ball SL, de Waard JR (2003b) Biological identifications through DNA barcodes. P Roy Soc Biol Sci 270:313–321. doi:10.1098/rspb.2002.2218
Hiddink JG, MacKenzie BR, Rijnsdorp A, Dulvy NK, Nielsen EE, Bekkevold D, Heino M, Lorance P, Ojaveer H (2008) Importance of fish biodiversity for the management of fisheries and ecosystems. Fish Res 90:6–8. doi:10.1016/j.fishres.2007.11.025
Hui D (2013) Global climate change and biodiversity: issues and future research. J Biodivers Endanger Species 1:e105. doi:10.4172/2332-2543.1000e105
Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA 86:6196–6200
Kris-Etherton PM, Harris WS, Appel LJ (2002) Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Circulation 106:2747–2757. doi:10.1161/01.CIR.0000038493.65177.94
Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. PNAS 105:2923–2928. doi:10.1073/pnas.0709936105
Madonna A, Alwany MA, Rabbito D, Trocchia S, Labar S, Abdel-Gawad FKh, D’Angelo R, Gallo A, Guerriero G, Ciarcia G (2015) Caves biodiversity in the marine area of Riviera d’Ulisse Regional Park, Italy: Grotta del Maresciallo overview. J Biodivers Endanger Species 3:153. doi:10.4172/2332-2543.10.4172/2332-2543.1000153
Mount DW (2007) Using a FASTA sequence database similarity search. Cold Spring Harb Protoc. doi:10.1101/pdb.top16
Mytilineou Ch, Akel EHKh, Babali N, Balistreri P, Bariche M, Boyaci YÖ, Cilenti L, Constantinou C, Crocetta F, Çelik M, Dereli H, Dounas C, Durucan F, Garrido A, Gerovasileiou V, Kapiris K, Kebapcioglu T, Kleitou P, Krystalas A, Lipej L, Maina I, Marakis P, Mavrič B, Moussa R, Peña-rivas L, Poursanidis D, Renda W, Rizkalla SI, Rosso A, Scirocco T, Sciuto F, Servello G, Tiralongo F, Yapici S, Zenetos A (2016) New Mediterranean biodiversity records (November, 2016). Medit Mar Sci 17:794–821. doi:10.12681/mms.1976
Occhipinti-Ambrogi A, Marchini A, Cantone G, Castelli A, Chimenz C, Cormaci M, Froglia C, Furnari G, Gambi MC, Giaccone G, Giangrande A, Gravili C, Mastrototaro F, Mazziotti C, Orsi-Relini L, Piraino S (2011) Alien species along the Italian coasts: an overview. Biol Invasions 13:215–237. doi:10.1007/s10530-010-9803-y
Patel DK (2014) Biodiversity and its Importance. J Biodivers Endanger Species 2:e117. doi:10.4172/2332-2543.1000e117
Puillandre N, Bouchet P, Boisselier-Dubayle MC, Brisset J, Buge B, Castelin M, Chagnoux S, Christophe T, Corbari L, Lambourdière J, Lozouet P, Marani G, Rivasseau A, Silva N, Terryn Y, Tillier S, Utge J, Samadi S (2012) New taxonomy and old collections: integrating DNA barcoding into the collection curation process. Mol Ecol Res 12:396–402. doi:10.1111/j.1755-0998.2011.03105.x
Rimbach G, Minihane AM, Majewicz J, Fischer A, Pallauf J, Virgli F, Weinberg PD (2002) Regulation of cell signalling by vitamin E. Proc Nutr Soc 61:415–425. doi:10.1079/PNS2002183
Scheffers BR, Joppa LN, Pimm SL, Laurance WF (2012) What we know and don’t know about Earth’s missing biodiversity. Trends Ecol Evol 27:501–510. doi:10.1016/j.tree.2012.05.008
Sievers F, Higgins DG (2013) Clustal Omega, accurate alignment of very large numbers of sequences. Method Mol Biol 1079:105–116. doi:10.1007/978-1-62703-646-7_6
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Trocchia S, Rabbito D, Angelo RD, Ciarcia G, Abdel-Gawad FKh (2015) Blue economy and biodiversity surveillance: fish caviar substitute rapid discrimination. J Biodivers Endanger Species 1:002. doi:10.4172/2332-2543.S1.002
Ward RD (2009) DNA barcode divergence among species and genera of birds and fishes. Molecular Ecology resources 8:1077–1085. doi:10.1111/j.1755-0998.2009.02541.x
Ward RD (2012) FISH-BOL, a case study for DNA barcodes. DNA Barcodes Methods Mol Biol 858:423–439. doi:10.1007/978-1-61779-591-6_21
Ward RD, Zemlak TS, Ines BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philos T Roy Soc B 360:1847–1857. doi:10.1098/rstb.2005.1716
Waugh J (2007) DNA barcoding in animal species: progress, potential and pitfalls. BioEssays 29:188–197. doi:10.1002/bies.20529
Acknowledgements
The authors thank the School of Nautical “Guardia di Finanza “of Gaeta, LT in Italy and the fishermen and the scuba divers in Egypt for granting the logistic organization in support of Adriano Madonna immersions. This work was supported by Federico II University, Naples, Italy.
Author information
Authors and Affiliations
Contributions
All authors contributed to conception and design of the experiments. All the authors have given their approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Animal rights
All applicable international, national, and/or institutional guidelines for welfare, care, and use of animals were followed.
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
Supported by a grant from Federico II University, Naples, Italy.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Guerriero, G., Rabbito, D., Alwany, M.A. et al. Fisheries and biodiversity along Mediterranean Sea: Italian and Egyptian coast overview. Euro-Mediterr J Environ Integr 2, 16 (2017). https://doi.org/10.1007/s41207-017-0027-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41207-017-0027-8