Abstract
MD2 pineapple production in the Valle del Cauca Department, Colombia, has increased in recent years due to its preference in the national and international markets. Pineapple mealybug wilt (PMW), reported in all production areas around the world, is associated with two mealybug species, Dysmicoccus brevipes and D. neobrevipes, and a group of virus species known as pineapple mealybug wilt-associated viruses (PMWaVs). In a previous study, D. brevipes was determined to be the main mealybug species associated with pineapples in the region; hence, the objective of this study was to determine the occurrence and distribution of PMWaVs in pineapple plants and specimens of D. brevipes collected in five MD2 pineapple fields in the Department of Valle del Cauca. Our results confirmed the presence of three virus species in both plant and mealybug specimens. PMWaV3 showed the highest detection frequencies (92.6% and 88.7%), followed by PMWaV1 (26.5% and 8.3%) and PMWaV2 (12.0% and 2.0%) in plant and mealybug samples, respectively. Mixed infections of PMWaVs were found also in the studied plant material. The most common virus association consisting of PMWaV1 and PMWaV3 was found in 30 plant samples (20%). The correlation analysis between each virus species found in mealybugs and their pineapple host suggests that PMWaV3 is not only transmitted through infested plant material, but that D. brevipes may also play a role in the dynamics of disease dissemination. The results of this study contribute to the understanding of PMWaV transmission in MD2 pineapple-cultivated areas in the Valle del Cauca region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since its discovery and dispersion, pineapple, Ananas comosus (L.) Merr. (Bromeliaceae), has been a popular and well-accepted fruit, becoming one of the most important tropical fruits worldwide (Reinhardt and Rodriguez 2009). In Colombia, the most common cultivars are India, Manzana, Mayanesa, MD2, and Perolera, each with their own organoleptic characteristics; however, the commercial MD2 hybrid is the preferred pineapple cultivar in the market as demonstrated by the increase of its cultivated area (Neira García et al. 2016; Rios 2020).
Pineapple mealybug wilt (PMW) is one of the most important diseases affecting pineapple production around the world (Rohrbach and Schmitt 2003; Hu et al. 2005; Perez et al. 2006). The disease is characterized by severe tip dieback, downward curving of leaf margins, reddening and wilting of leaves, and can cause total collapse of the plant (Sether and Hu 2002a, b; Jahn et al. 2003; Dey et al. 2018). PMW is known to be associated with two species of mealybugs, i.e., the pink pineapple mealybug, Dysmicoccus brevipes (Cockerell), and the gray pineapple mealybug, D. neobrevipes Beardsley (Hemiptera: Coccomorpha: Pseudococcidae) (Carter 1963; Sartiami and Kondo 2022). PMW was originally thought to be a result of phytotoxicity that occurred during the insect feeding process (Carter and Collins 1933). However, the presence of asymptomatic plants which were heavily infested with mealybugs suggested an association with a transmissible factor (Ito 1959). For many years, the causative agent of PMW remained unknown. However, in 1989, double-stranded RNA, together with flexible particles and a capsid protein of around 23.8 KDa, was detected in symptomatic pineapple plants which were absent in healthy plants (Gunasinghe and German 1989). Based on their characteristics, the particles were assigned to the genus Ampelovirus and referred to as pineapple mealybug wilt-associated viruses (PMWaVs) (Agranovsky 1996). Studies conducted in Hawaii and Australia identified three virus species named as PMWaV1, PMWaV2, and PMWaV3 and a putative species named as PMWaV5 associated with PMW (Melzer et al. 2001, 2008; Gambley et al. 2008; Sether et al. 2009). A virus previously designated as PMWaV4 now appears to be a strain of PMWaV1 (Green et al. 2020) and PMWaV6 has been recently proposed as a putative new member of the genus Ampelovirus (Larrea-Sarmiento et al. 2021). Phylogenetic analyses of the above virus species have classified PMWaV1, PMWaV3, and the putative species PMWaV5 in the subgroup II of the genus Ampelovirus and PMWaV2 and the putative species PMWaV6 in the subgroup I (Dey et al. 2018; Larrea-Sarmiento et al. 2021). The development of diagnostic techniques for each virus species has made possible the validation of the presence of PMWaVs in plant material as well as their transmission by an insect vector (Sether et al. 1998, 2005; Sether and Hu 2002b; Gambley et al. 2008). Different studies have confirmed that the three currently classified PMWaV species by the International Committee on Taxonomy of Viruses (ICTV) PMWaV1, PMWaV2, and PMWaV3 are widely distributed in different pineapple production areas around the world (Sether et al. 2005; Gambley et al. 2008; Asare-Bediako et al. 2020; Hernandez-Rodriguez et al. 2014; Ochoa-Martinez et al. 2016; Massé et al. 2021). However, a clear association between PMWaV2 and PMW in the presence of mealybug feeding has only been reported in Hawaii (Sether and Hu 2002a). Furthermore, in Cuba, Hernandez-Rodriguez et al. (2014) reported a wide distribution of these three virus species in commercial pineapple cultivation areas and a high occurrence of PMW symptoms, which they suggested to be associated with the high detection frequency of PMWaV2. In Australia, PMWaV1, PMWaV2, PMWaV3, and the putative species PMWaV5 have been found in symptomatic and asymptomatic pineapple plants without a clear correlation with PMW in the studied area (Gambley et al. 2008). So far in Colombia, there are no studies on the distribution of PMWaVs and their association with PMW in commercial cultivation areas. A previous study detected PMWaV2 in two departments, namely the Department of Meta, municipality of San Martin, in Smooth Cayenne and the MD2 pineapple hybrid, and in the Department of Quindío, municipality of Montenegro, in the Manzana cultivar (Rodelo 2007, as cited in Moreno et al. 2021). Furthermore, a recent study reported the pink pineapple mealybug, D. brevipes, as the only mealybug species found on pineapples in the MD2 pineapple-cultivated area of the Department of Valle del Cauca (Moreno et al. 2021).
Considering the importance of MD2 pineapple cultivation in the Valle del Cauca region, the confirmed prevalence of D. brevipes, and its possible role as a vector of PMWaVs, the objective of this study was to determine the occurrence and distribution of PMWaV1, PMWaV2, and PMWaV3 in pineapple plants and mealybugs. This study should contribute to the understanding of the mechanism of virus transmission in the region, required for the development of integrated pest management strategies.
Materials and methods
Sample collection
Plant and insect material (i.e., mealybugs) was collected from pineapple fields in five municipalities (i.e., Dagua, La Cumbre, Restrepo and Vijes located in the upper land zone, and Palmira located in the low land zone) in the Department of Valle del Cauca, Colombia, during the period from May to August 2019 as reported by Moreno et al. (2021). The study was carried out in growers’ pineapple fields between 1 and 4 ha in the second cycle of production, cultivated for the local market. All sampled plants were in the reproductive growth phase at stage 7 (fruit development) or stage 8 (fruit ripening) according to the phenological growth stages of pineapple described by Zhang et al. (2016). Fields were selected based on two criteria, i.e., (i) they were planted with MD2 pineapple cultivar, and (ii) had populations of D. brevipes. It should be noted that pineapples of the MD2 cultivar in the explored fields did not manifest PMW symptoms. Sample collections were conducted under a collecting permit by the Colombian National Authority for Environmental Permits (resolution nº. 1466 - December 3, 2014). In each selected field where the presence of D. brevipes was previously confirmed (Moreno et al. 2021), a random exploration within the plots was conducted, selecting 30 plant samples based on the presence of mealybugs. The collected plant material consisted of young well-formed leaves which were cleaned with 70% ethanol and dried with a paper towel, then put into a paper bag, sealed in a plastic bag, and labelled with a serial number and collecting data. From the same plants, mealybug-infested roots were stored in a plastic bag and transported to the entomology laboratory at Agrosavia, Palmira Research Station, where the adult female mealybugs were collected with a paint brush and stored in vials filled with 70% and 90% ethanol for subsequent morphological identification and RNA extraction, respectively. Details on collecting and preserving mealybug specimens can be found in Kondo and Watson (2022).
Molecular detection of PMWaV1, PMWaV2, and PMWaV3 from plant and insect material
Sample preparation and RNA extraction
Samples were processed on their arrival at the laboratory to allow for proper storage. From each collected plant sample, the tissue close to the basal part of the leaves was cut into small pieces with a sterilized pair of scissors. The pieces of plant tissue were ground with liquid nitrogen until becoming a fine powder and then transferred to 2-ml Eppendorf tubes and stored at −80 °C. The RNA extraction protocol was standardized based on the protocol reported by Chang et al. (1993). For each sample, an amount between 250 and 400 mg of powdered tissue was processed and the recovered RNA was suspended in 30 µl of DEPC water. The integrity of the nucleic acid was verified by 2% W/V agarose gel electrophoresis and the RNA concentration was estimated by spectrometry using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA). Samples were kept at −80 °C.
The adult female insects that had been collected from the roots and put in vials filled with 90% ethanol were stored at −20 °C. Mealybug RNA was extracted from three or more mealybugs measuring between 1 and 3 mm in length to recover a sufficient RNA concentration for the analyses. The RNA extraction for the mealybugs was performed using the RNeasy Plant Mini Kit (Qiagen) with slight modifications for the initial steps of sample preparation, i.e., mealybugs were ground directly with an aliquot of RLT buffer using a sterile plastic pestle in an Eppendorf tube, and then, the other aliquot of RLT buffer with 0.1% 2-mercaptoethanol was added to complete the volume required and vortexed. The tube was briefly spun to collect the remains of mealybug exuviae and undissolved detritus before transferring to a QiaShredder column. Thereafter, RNA extraction was performed following the instructions of the provider. The recovered RNA was eluted in 30 µl of DEPC water. The RNA integrity and concentration was verified as for the plant material.
Amplification by RT-PCR of PMWaV1, PMWaV2, and PMWaV3 specific region
Complementary DNA (cDNA) of plant and insect material was synthetized from 200 ng of RNA template. For plant and insect material, the Verso Kit (Thermo Fisher Scientific, Waltham, USA) was used. The specific reverse primers for each virus species plus random hexamers were included in the reaction following the instructions of the commercial provider. cDNA samples were stored at −20 °C until processing. The detection of PMWaV1, PMWaV2, and PMWaV3 in the collected samples was conducted through the amplification by reverse transcription polymerase chain reaction (RT-PCR) of the conserved region of the Ampelovirus genome. The RT-PCR reaction was conducted with the previously reported set of primer pairs V1-225/V1-226, V2-224/V2-223, and V3-263/V3-264 for PMWaV1, PMWaV2, and PMWaV3, respectively (Sether et al. 2001, 2005) (Table 1). The primer pairs were designed to amplify a specific region of the heat shock protein 70 homolog (HSP70h) of 589, 609, and 495 bp for PMWaV1, PMWaV2, and PMWaV3, respectively. Each RT-PCR reaction included a positive control (+ C) corresponding to the protein target region for each species cloned into a commercial plasmid, which was kindly provided by the Analysis and Diagnostic Laboratory of the Colombian Agricultural and Livestock Institute-ICA (Spanish acronym for Instituto Colombiano Agropecuario) (Mosquera, Cundinamarca), and a control for the reaction consisting of ultrapure water instead of cDNA template (−C). The amplification reaction was standardized under laboratory conditions using MyTaq Red Mix 2X (Meridian Bioscience, Cincinnati, USA); for each reaction, 0.84X My Taq Red Mix 2X, 0.2 µM of each primer, and 2 µl of cDNA were added for master mix preparation in a final volume of 25 µl. The amplification program was run in a SureCycler 8800 (Agilent Technologies, Santa Clara, USA) and consisted of an initial denaturation step at 95 °C for 3 min, followed by 35 cycles of an initial step at 95 °C for 30 s, 55 °C (V1-224/V1-226, V2-223/V2-224, and V3G3-F/V3G3-R) or 60 °C (V3-263/V3-264) for 60 s, 72 °C for 60 s, and a final elongation at 72 °C for 10 min.
In addition, a new pair of specific primers for PMWaV3 detection in both plant and insect materials was manually designed based on a conserved region of HSP70h gene. A specific PMWaV3 region within the HSP70h genes was determined by the alignment of MH704740.1 (PMWaV1), MH704741.1 (PMWaV2), and MH704742.1 (PMWaV3) accessions using ClustalW tool of MEGA X software (Kumar et al. 2018). Primers flanking the region selected above were designed and primer-primer interactions were tested with the AutoDimer software (Vallone and Butler 2004). The new pair of primers V3G3-F and V3G3-R was located at positions 8395 and 8690 of PMWaV3 viral genome (MH704742.1), corresponding to a product size of 296 bp within the HSP70h gene. The RT-PCR amplification conditions were the same as for the other pairs of primers. The amplified products were visualized by electrophoresis in 1.6% (W/V) agarose gel dyed with GelRed (Biotium, Fremont, USA) using the gel documentation system Enduro GDS (Labnet, Edison, USA). Samples with presence of the expected band were re-amplified in order to confirm the results before being recorded as a positive result.
In order to confirm the molecular identity of the HSP70h region in PMWaV1, PMWaV2, and PMWaV3, a group of samples with the expected bands (Sether et al. 2001, 2005) were sequenced. Bands from the amplified products obtained by RT-PCR were eluted directly from gel using a scalpel and purified with the QIAquick PCR and Gel Cleanup kit (Qiagen, Hilden, Germany). Samples were sent for direct amplicon sequencing to Macrogen (Seoul, Korea). Amplified products from mealybugs were shipped with the export license no. 02865 issued by the Colombian National Authority for Environmental Permits. The forward and reverse sequences obtained were edited and aligned using the software Geneious Prime 2021.0.3. Lastly, the sequence identities were analyzed using the Blastn tool at the National Center for Biotechnology Information (NCBI). Sequences showed good quality but only the consensus sequences from each virus species were deposited in GenBank (NCBI).
Data analyses
The mealybug-infested plants from which plant tissue and insect material were collected were defined as the experimental unit. The results obtained from the molecular detection study of the three virus species on each plant (plant tissue and insect material) were recorded into a binary matrix (0: Absence and 1: Presence) for the analyses. First, the detection frequency of each virus species (i.e., PMWaV1, PMWaV2, and PMWaV3) in both plant and insect materials was calculated with the formula Df = a/b×100 where a is the number of plant or insect samples with the presence of each virus species and b is the total number of analyzed samples (plant or insect), and the distribution of the occurrence of each virus species per municipality was determined by a table of frequencies. Second, the molecular detection of the virus species in plant material was visualized by network modeling of the results based on the presence of the virus species using different colors, PMWaV1 in red, PMWaV2 in blue, and PMWaV3 in green, using the Cytoscape Software (Shannon et al. 2003). Third, in order to test a possible relationship between the two variables, that is, (i) the presence of each virus species in the mealybug and (ii) its further presence in the plant material, a statistical analysis of dichotomic qualitative variables using contingency tables was conducted. A chi square (X2) with a critical value (X 2(1;0,05) = 3,841) was applied for each relationship to accept or reject the null hypothesis of independence between the two analyzed variables. When a dependence between variables was found, the V of Cramer statistics was applied to determine the intensity of the association. V Cramer’s values close to 0 indicate a weak association and close to 1 means a strong association. The analyses were conducted with the R statistics program, version 4.0.3 (R Core Team 2021).
Results
Molecular detection of PMWaV1, PMWaV2, and PMWaV3 from plant and insect material
For the molecular detection of the virus species, a total of 150 samples from plant tissue (i.e., leaves) and 142 mealybug samples were collected and processed. The reduced number of mealybugs in the analysis was due to the loss during transportation of mealybugs that were collected from the roots for RNA extraction. The RNA samples extracted from plant and mealybug samples were characterized by good quality and high concentrations. The RNA concentrations from plant material ranged between 496.5 and 3101 ng/µl, and in insect material, the concentrations ranged between 29.7 and 892.9 ng/µl, with differences being associated with the amount and size of individuals used for the extraction.
Amplification by RT-PCR of PMWaV1, PMWaV2, and PMWaV3 in plant and insect material collected in this study allowed the identification of specific viruses with expected bands of 589 bp (PMWaV1), 609 bp (PMWaV2) (Sether et al. 2001), and 495 bp (PMWaV3) (Sether et al. 2005), respectively, with previously reported primers, and an expected band of 296 bp with the PMWaV3 primers developed in this study. From the 150 plant samples and 142 mealybugs that were analyzed, the following number of samples showed the expected band: for PMWaV1, 40 plant and 12 mealybug samples, respectively (Fig. 1A and B); for PMWaV2, 18 plant and 3 mealybug samples, respectively (Fig. 1C and D); for PMWaV3 with the primer set V3-263/V3-264, 121 plant and 0 mealybug samples, respectively (Fig. 1G); and with the primer set V3G3-F/V3G3-R, 139 plant and 126 mealybug samples, respectively (Fig. 1E and F). Considering the specificity of the primers and the results found for the mealybugs, the data obtained with the pair of PMWaV3 primers developed in this study was used for the analyses.
Agarose gel electrophoresis of RT-PCR amplified products using V1-225/V1-226 primers in plant (A) and insect samples (B) for PMWaV1 detection; V2-224/V2-223 in plant (C) and mealybug samples (D) for PMWaV2 detection; V3G3-F/V3G3-R in plant (E) and mealybug samples (F) and V3-263/V3-264 in plant material (G) for PMWaV3 detection. MW, molecular weight marker 100 bp; MW*, Easy Ladder I (Meridian Bioscience Cincinnati, USA) showing bands of 100, 200, 500, 1000, and 2000 bp; lanes 1–42 and lanes 43–66 correspond to random plant and mealybug samples respectively used for PMWaV detection; −C, no template control; +C, positive control. White arrows in some samples indicate amplified fragments when unspecific bands were present, and black arrows indicate the expected products’ size
The analysis of sequences from the amplified products for each virus species allowed the confirmation of their sequence identities with those reported in GenBank (NCBI). From each group, the accession number of the consensus sequences deposited in GenBank is described in parentheses. For PMWaV1, 11 samples from plant material (ON988055, ON988057, ON988060, ON988063, ON988067, and ON988074 to ON988076) and six from insect material (OP153792 and OP153794) shared identity of 98.14% with the GenBank accession HQ129930.1, corresponding to a sequence from Cuba. For PMWaV2, 11 samples from plant material (ON988061, ON988064, ON988068, ON988072, and ON988077) and two from insect material (OP153795 and OP153796) showed identities between 99.31 and 100% with GenBank accession KT322167.1, corresponding to a sequence from Thailand. Finally, for PMWaV3 with the primer set V3-263/V3-264, eight samples from plant material (ON988056, ON988062, ON988065, ON988070, ON988071, and ON988073) showed an identity over 97.98% with GenBank accession JX508636.1, corresponding to a sequence from Cuba. With the pair of primers developed in this study (V3G3-F/V3G3-R), six samples from plant material (ON988053, ON988054, ON988058, ON988059, ON988066, and ON988069) and five of mealybugs (OP153788 to OP153791 and OP153793) showed identities above 97.64% with GenBank accession MN539274.1, corresponding to PMWaV3 genome from Hawaii.
Data analyses
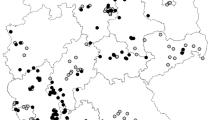
Regarding the first analysis, for PMWaV1, 40 plant samples showed presence of the virus, with a detection frequency of 26.5% (40/150) in the study region. PMWaV1 was locally distributed in Dagua and Vijes with values of 46.7% (14/30) and 36.7% (11/30), respectively, followed by Palmira 26.7% (8/30), Restrepo 16.7% (5/30), and La Cumbre 6.7% (2/30) (Fig. 2A). The detection frequency of PMWaV1 in the mealybug population was 8.4% (12/142) distributed in Vijes 16% (4/25) and Dagua 14.3% (4/28), followed by Palmira 10.0% (3/30) and La Cumbre 3.3% (1/30) (Fig. 2B).
For PMWaV2, the virus was detected in 18 plant samples, with the lowest detection frequency in the population of 12.0% (18/150), distributed in the localities of Vijes 30.0% (9/30), Dagua 20.0% (6/30), Palmira 6.7% (2/30), and Restrepo 3.3% (1/30) (Fig. 2A). The detection frequency of PMWaV2 in the mealybug population was 2.0% (3/142) and only detected in Vijes 12.0% (3/25) (Fig. 2B).
Using the primer set V3G3-F/V3G3-R, 139 samples of plant material showed the expected region for PMWaV3, which corresponds to a detection frequency of this virus in the population of 92.6% (139/150). The distribution of PMWaV3 in pineapple plants in all five localities was uniform, showing values of 96.7% (29/30), 96.7% (29/30), 93.3% (28/30), 90.0% (27/30), and 86.7% (26/30) in Vijes, Palmira, Restrepo, La Cumbre, and Dagua, respectively (Fig. 2A). The detection frequency of PMWaV3 in the mealybug population was 88.7% (126/142), and showed a uniform distribution in all localities: Palmira 90.0% (27/30), followed by Restrepo with 89.7% (26/29), Vijes 100.0% (25/25), La Cumbre 83.3% (25/30), and Dagua with 82.1% (23/28) (Fig. 2B).
The network modeling analysis allowed the visualization of the distribution of each virus species among the positive plant samples, and mixed infections by two or three virus species (Fig. 3). The most common multiple infections were PMWaV1/PMWaV3 and found in 30 samples (20%), followed by PMWaV1/PMWaV2/PMWaV3 in 9 samples (6%), PMWaV2/PMWaV3 in 8 samples (5%), and PMWaV1/PMWaV2 in 1 sample (0.6%); single infections by PMWaV3 were recorded in 92 samples (61%) (Fig. 3).
For the correlation analysis, only the data from 142 plants with results for both plant and mealybug materials were analyzed. The null hypothesis states that the variables, i.e., presence of PMWaVs in mealybugs and their further presence in plants, are independent (i.e., they have no association), against the alternative hypothesis that the variables are not independent; in other words, the presence of PMWaVs in mealybugs and in their corresponding host plants may be associated with vector transmission. Based on our analyses, the null hypothesis of independence of the variables was rejected for PMWaV1, PMWaV2, and PMWaV3 with a X2 = 26.751, X2 = 5.0452, and X2 = 67.256, respectively. Hence, it was assumed that a level of dependence exists between the presence of each virus species in mealybugs and pineapple plants, but the intensity of the association based on the V of Cramer was different: for PMWaV1, the V of Cramer (0.463) ≈ 0.5 did not show a clear intensity of the association; for PMWaV2, the V of Cramer (0.268) ≈ 0.3 showed a weak association between the two variables; and for PMWaV3, the V of Cramer (0.73) ≈ 0.7 showed a strong association between the two variables (Fig. 4). In summary, the correlation analysis indicates a strong association between the presence of PMWaV3 in mealybugs and plant material.
Discussion
Herein, we report the detection of the PMWaVs in asymptomatic MD2 pineapple plants and for the first time in D. brevipes in five localities of the Valle del Cauca Department, Colombia. The highest detection frequencies in plants and mealybugs were found for PMWaV3 (92.6% and 88.7%, respectively), followed by PMWaV1 (26.5% and 8.3%, respectively) and PMWaV2 (12.0% and 2.0%, respectively). PMWaV3 was also characterized by having a uniform distribution in the studied localities in both plants and mealybugs. Similar results have been reported in Australia, where PMWaV1 and PMWaV3 were found to be more widespread with a high detection frequency for PMWaV3 of 67.5%, whereas PMWaV2 was detected in less than 25% of tested plants (Gambley et al. 2008). Moreover, similar PMWaV1 frequency values were reported by Sether et al. (2010) from different localities in Hawaii which ranged between 15.0 and 22.8% in plant material. In another study, a frequency of PMWaV1 detection of 20% was found in in vitro plants propagated from asymptomatic MD2 pineapple plants collected in the field in Cuba (Hernández-Rodríguez et al. 2017). Contrary to our results, in Cuba, Ghana, and Hawaii, PMWaV2 has been reported as having the highest frequencies and in Hawaii this virus species was found directly associated with PMW (Hernández-Rodríguez et al. 2017; Dey et al. 2018; Asare-Bediako et al. 2020).
Our results suggest a longer and more frequent exposure of MD2 plants to PMWaV3 than to the other viruses in the Valle del Cauca region. PMWaV3 is the only virus species present in multiple and single infections. The high frequencies of PMWaV3 in the region could be related to an evolutionary advantage of this species associated with the combined effect of its early invasion, the common use of suckers from the same plants for plant propagation, and the presence of its potential vector, D. brevipes. However, a current association between the PMWaV species found and PMW could not be confirmed because of the absence of symptoms in the explored MD2 pineapple fields. In Colombia, PMW symptoms have been previously reported associated with PMWaV2 infection in two pineapple cultivars in non-commercial fields in two municipalities: San Martin, Department of Meta in cv. Smooth Cayenne and MD2 hybrid, and in Montenegro, Department of Quindío, in cv. Manzana (Rodelo 2007, as cited in Moreno et al. 2021). However, we could not find an association of PMW symptoms and PMWaV2 infection in commercial MD2 pineapple fields in the Valle del Cauca region. This could be associated with the low detection frequency of PMWaV2 in the studied material of this cultivar. Moreover, the impact of PMWaV infection on MD2 pineapple production and fruit quality has not been noticed by growers because infected plants show no visible symptoms. However, pineapple growers need to consider the risk of using their own plant material for replanting the following production cycles or of exchanging plant material between neighbors. The continued use of infected material could favor mutations in the virus species and the appearance of new virus strains and species with an unpredictable impact on production.
Additionally, the results of this study on data correlation suggest that besides the transmission of PMWaVs through infested plant material, the pineapple mealybug, D. brevipes, may also be playing a role in virus transmission; however, further studies are needed to confirm this hypothesis. The ability of D. brevipes to acquire and transmit the three PMWaV species has been determined in previous studies (Sether et al. 1998, 2005; Sether and Hu 2002b; Gambley et al. 2008). Transmission of PMWaVs by mealybugs is considered semi-persistent (Sether et al. 1998) and the level of vector-pathogen specificity for different virus species varies in the family Closteroviridae; some viruses have a wide vector range and others have been demonstrated to be transmitted by only one insect species (Karasev 2000). More studies are needed in order to determine the efficiency of virus transmission in different pineapple cultivars and the contribution of D. brevipes as a vector of PMWaV3 in the region.
In conclusion, our results confirmed the presence of three virus species, i.e., PMWaV1, PMWaV2, and PMWaV3, in plant material and for the first time in D. brevipes in the Department of Valle del Cauca, Colombia. At the same time, we report a high detection frequency of PMWaV3 in both pineapple plants and mealybugs, with uniform distribution at the studied localities. These findings will contribute to the understanding of PMWaV transmission in the region, which is required for the development of integrated pest management strategies.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Agranovsky AA (1996) Principles of molecular organization, expression, and evolution of closteroviruses: over the barriers. Advances in Virus Research 47:119–158
Asare-Bediako E, Nyarko J, Puije GC (2020) First report of pineapple mealybug wilt associated virus‐2 infecting pineapple in Ghana. New Disease Reports 41:9–9
Carter W (1963) Mealybug wilt of pineapple; a reappraisal. Annals of the New York Academy of Sciences 105:741–764
Carter W, Collins JL (1933) The pineapple mealy bug, Pseudococcus brevipes (ckl.) And wilt of pineapples. Phytopathology 23:207–242
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Report 11:113–116
Dey KK, Green JC, Melzer MJ, Borth W, Hu JS (2018) Mealybug wilt of pineapple and associated viruses. Horticulturae 4:52
Gambley CF, Steele V, Geering AD, Thomas JE (2008) The genetic diversity of ampeloviruses in australian pineapples and their association with mealybug wilt disease. Australian Plant Pathology 37:95–10
Green JC, Rwahnih MA, Olmedo-Velarde A, Melzer MJ, Hamim I, Borth WB, Brower TM, Wall M, Hu JS (2020) Further genomic characterization of pineapple mealybug wilt-associated viruses using high-throughput sequencing. Tropical Plant Pathology 45:64–72
Gunasinghe UB, German TL (1989) Purification and partial characterization of a virus from pineapple. Phytopathology 79:1337–1341
Hernández-Rodríguez L, Nápoles L, Pérez L, Concepción O, Cid M, Alvares Y, Zamora V (2017) Infección de pineapple mealybug wilt-associated virus 1, 2 y 3 en plantas de piña, híbrido ‘MD-2’ en ciego de Ávila. Centro Agricola 44:52–60
Hernandez-Rodriguez L, Ramos-Gonzalez PL, Garcia-Garcia G, Zamora V, Peralta-Martin AM, Peña I, Perez JM, Ferriol X (2014) Geographic distribution of mealybug wilt disease of pineapple and genetic diversity of viruses infecting pineapple in Cuba. Crop Protection 65:43–50
Hu JS, Sether DM, Metzer MJ, Pérez E, Gonsalves A, Karasev AV (2005) Pineapple mealybug wilt associated virus and mealybug wilt of pineapple. Acta Horticulturae 666:209–212
Ito K (1959) Terminal mottle as a symptomatological aspect of mealybug wilt with evidence supporting the hypothesis of a virus etiology of the disease. Research report (Pineapple Research Institute of Hawaii), Honolulu
Jahn GC, Beardsley JW, González-Hernández H (2003) A review of the association of ants with mealybug wilt disease of pineapple. Proceedings of the Hawaiian Entomological Society 36:9–28
Karasev AV (2000) Genetic diversity and evolution of closteroviruses. Annual Reviews of Phytopathology 38:293–324
Kondo T, Watson GW (2022) Chap. 5. Collection, Preservation, Slide mounting, labelling and vouchering of Scale Insects. In: Kondo T, Watson GW (eds) Encyclopedia of scale insect pests. CABI International, Wallingford, pp 548–558
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35:1547–1549
Larrea-Sarmiento A, Olmedo-Velarde A, Wang X, Borth W, Matsumoto TK, Suzuki JY, Wall MM, Melzer M, Hu J (2021) A novel ampelovirus associated with mealybug wilt of pineapple (Ananas comosus). Virus Genes 57:464–468
Massé D, Cassam N, Hostachy B, Iskra-Caruana ML, Darnaudery M, Lefeuvre P, Lett JM (2021) First report of three pineapple mealybug wilt-associated viruses in queen victoria pineapples in Reunion Island. Plant Disease 105:715
Melzer MJ, Karasev AV, Sether DM, Hu JS (2001) Nucleotide sequence, genome organization and phylogenetic analysis of pineapple mealybug wilt-associated virus-2. The Journal of General Virology 82:1–7
Melzer MJ, Sether DM, Karasev AV (2008) Complete nucleotide sequence and genome organization of pineapple mealybug wilt-associated virus-1. Archives of Virology 153:707–714
Moreno I, Tarazona-Velásquez R, Campos-Patiño Y, Rodríguez-Arévalo KA, Kondo T (2021) Prevalence of Dysmicoccus brevipes (Cockerell, 1893) (Hemiptera: Pseudococcidae) in MD2 pineapple crop areas in Colombia. Pesquisa Agropecuaria Tropical 51:e67838
Neira García AM, Martínez Reina AM, Rodríguez Orduz JO (2016) Análisis del mercado de piña gold y perolera en dos principales centrales mayoristas de Colombia. Corpoica Ciencia y Tecnología Agropecuaria 17:149–165
Ochoa-Martínez DL, Uriza-Ávila DE, Rojas-Martínez RI (2016) Detection of pineapple mealybug wilt-associated virus 1 and 3 in Mexico. Revista Mexicana de Fitopatología 34:131–141
Perez EP, Sether DM, Melzer MJ, Busto JL, Hu JS (2006) Characterization and control of pineapple mealybug wilt associated ampeloviruses. Acta Horticulturae 702:23–27
R Core Team (2021) R: A language and environment for statistical computing, Vienna. Available at: https://www.R-project.org/. Accessed 1 Sept 2021
Reinhardt A, Rodriguez LV (2009) Industrial processing of pineapple – trends and perspectives. VI International Pineapple Symposium 822:323–328
Rios L (2020) Colombia es un país rico en variedades locales de piña. Red Agricola, Frutales 6:36–38. Available at: https://www.redagricola.com/co/assets/uploads/2020/11/racol06.pdf. Accessed 18 Oct 2021
Rohrbach KG, Schmitt D (2003) Diseases of pineapple. In: Ploetz R (ed) Diseases of tropical fruit crops. CAB International, Cambridge, pp 443–464
Sartiami D, Kondo T (2022) Dysmicoccus spp. (Hemiptera: Coccomorpha: Pseudococcidae). In: Kondo T, Watson G (Eds.) Encyclopedia of scale insect pests CABI International, Wallingford, pp 162–177
Sether DM, Borth WB, Melzer MJ, Hu JS (2010) Spatial and temporal incidences of pineapple mealybug wilt-associated viruses in pineapple planting blocks. Plant Disease 94:196–200
Sether DM, Hu JS (2002) Yield impact and spread of pineapple mealybug wilt associated virus-2 and mealybug wilt of pineapple in Hawaii. Plant Disease 86:867–874
Sether DM, Hu JS (2002) Closterovirus infection and mealybug exposure are necessary for the development of mealybug wilt of pineapple disease. Phytopathology 92:928–935
Sether DM, Karasev AV, Arakawa C, Zee F, Kislan MM, Busto JL, Hu JS (2001) Differentiation, distribution, and elimination of two different pineapple mealybug wilt-associated viruses found in pineapple. Plant Disease 85:856–864
Sether DM, Melzer MJ, Borth W, Hu JS (2009) Genome organization and phylogenetic relationship of pineapple mealybug wilt associated virus-3 with family Closteroviridae members. Virus Genes 38:414–420
Sether DM, Melzer MJ, Busto J, Zee F, Hu JS (2005) Diversity and mealybug transmissibility of ampeloviruses in pineapple. Plant Disease 89:856–864
Sether DM, Ullman DE, Hu JS (1998) Transmission of pineapple mealybug wilt-associated virus by two species of mealybug (Dysmicoccus spp.). Phytopathology 88:1224–1230
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13:2498–2504
Vallone PM, Butler JM (2004) AutoDimer: a screening tool for primer-dimer and hairpin structures. BioTechniques 37:220–231
Zhang HN, Sun WS, Sun GM, Liu SH, Li YH, Wu QS, Wei YZ (2016) Phenological growth stages of pineapple (Ananas comosus) according to the extended Biologische Bundesantalt, Bundessortenamt and Chemische Industrie scale. The Annals of Applied Biology 169:311–318
Acknowledgements
The authors are grateful to Dr. Penny J. Gullan (Australian National University, Canberra, Australia) and Dr. Yamila Rodríguez (National Center for Animal and Plant Health, CENSA, Cuba) for kindly reviewing the manuscript. Thanks also to Yenifer Campos-Patiño, Juliana Moncada and Hebert Hernández for their assistance in the collection and preparation of samples.
Funding
Open Access funding provided by Colombia Consortium. This research work is part of the results of a project entitled “Technological options for the development and sustainable management of pineapple cultivation Ananas comosus (L.) Merr. (Bromeliaceae) in the main producing areas of Colombia,” funded by the Ministry of Agriculture and Rural Development (Ministerio de Agricultura y Desarrollo Rural de Colombia).
Author information
Authors and Affiliations
Contributions
IM conceived and designed the work, interpreted data, and prepared the first draft of the manuscript; KAR-A performed the lab experiments and generated and interpreted data. RT-V conducted the data analyses; TK interpreted data and wrote and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Moreno, I., Rodríguez-Arévalo, K.A., Tarazona-Velásquez, R. et al. Occurrence and distribution of pineapple mealybug wilt-associated viruses (PMWaVs) in MD2 pineapple fields in the Valle del Cauca Department, Colombia. Trop. plant pathol. 48, 217–225 (2023). https://doi.org/10.1007/s40858-023-00559-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-023-00559-8