Abstract
Purpose
To improve the performance of deep-learning-based image segmentation, a sufficient amount of training data is required. However, it is more difficult to obtain training images and segmentation masks for medical images than for general images. In deep-learning-based colon polyp detection and segmentation, research has recently been conducted to improve performance by generating polyp images using a generative model, and then adding them to training data.
Methods
We propose SemanticPolypGAN for generating colonoscopic polyp images. The proposed model can generate images using only the polyp and corresponding mask images without additional preparation of input condition. In addition, the semantic generation of the shape and texture of polyps and non-polyp parts is possible. We experimentally compare the performance of various polyp-segmentation models by integrating the generated images and masks into the training data.
Results
The experimental results show improved overall performance for all models and previous work.
Conclusion
This study demonstrates that using polyp images generated by SemanticPolypGAN as additional training data can improve polyp segmentation performance. Unlike existing methods, SemanticPolypGAN can independently control polyp and non-polyp parts in a generation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
According to the International Agency for Research on Cancer, colorectal cancer is the third most common type of cancer worldwide and has the second highest mortality rate [1]; the 5 year relative survival rate for colorectal cancer from 2013 to 2019 was 65% [2]. Colon cancer can be prevented if polyps are detected and removed early [3]. One of the ways to detect polyps is through colonoscopy. However, the rate of missing polyps during colonoscopy varies from 6 to 27% [4].
Recent studies on colon polyp detection [5,6,7] and segmentation [8,9,10,11,12] have used deep learning. However, medical data such as colon polyp images, are more difficult to collect than general images. Due to privacy, personal medical data cannot be fully utilized [13]. Even with sufficient data, skilled experts are needed to label polyp masks for annotation, consuming significant time and costs. Therefore, most polyp studies use publicly available data for research purposes [14,15,16,17]. Due to limited data, the diversity of polyps is insufficient, limiting the performance of deep-learning models. To overcome these limitations, studies are being conducted on generating various synthetic colon polyp images for use as deep-learning training data for polyp detection and segmentation to improve performance [18,19,20].
In [18], the generator of pix2pix [21] model was modified to generate polyp images using polyp mask images as input. The authors augmented training images by generating additional images as training data and achieved improved polyp detection and segmentation performance. However, the model cannot generate images for normal parts without polyps, and the characteristics of the generated polyps are limited to the training images.
In [19], a conditional generative adversarial network (GAN) [22] was used to generate polyp images. To generate realistic polyp images, edge filtering was applied to the polyp image. Thereafter, the location of the polyp mask was indicated on the edge filtering image, and used as a condition image. In the inference phase, edge filtering was used for normal colon images, and an arbitrary polyp mask was synthesized thereon and used as input. A conditional image preparation step and a normal colonoscopy image without polyps are required as input. Additionally, it is difficult to generate polyps of various characteristics.
In [20], the goal was to generate synthetic polyp images using only the provided polyp dataset, without additional preparation such as a separate normal dataset and edge filtering. Different labels were manually inserted into the polyp part of the generated polyps with the desired characteristics. The generated images were additionally used as training images for the polyp object-detection and segmentation model, improving performance. However, due to limited training data, the process requires transforming polyp images into normal images and then reversing them back into polyp images. Images must be labeled manually to control polyp characteristics; however, it is impossible to control the shape and characteristics of non-polyp parts.
StyleGAN [23] is used to combine the styles of general images. The image is considered a combination of several styles and is composited by applying style information each time through each layer. However, it is impossible to control each class independently; all classes are controlled at once.
Unlike [23], SemanticStyleGAN [24] can independently control style and semantic elements. It can also control the shape and texture of each element. The authors used face data with fixed elements as an input mask to control each of them. A method was used to create a generator for each element and generate them independently rather than all at once and then synthesize them. This enabled combining generated face images or transforming only desired parts of a specific image, such as the eyes, nose, and mouth.
Based on SemanticStyleGAN [24], we propose SemanticPolypGAN, which can control the shape and texture of polyps and non-polyp parts while generating polyps. Unlike existing polyp-generation methods, it is possible to generate polyp images and polyp masks without additional input preparation steps. The shape and texture can be controlled by randomly modifying the latent vector of the generated polyp image. Semantic synthesis between generated polyp images is also possible. We explore polyp-segmentation performance improvement by adding the generated polyp images and masks to training data. To evaluate segmentation performance, polyp segmentation models UACANet [8], PraNet [9], TGANet [10], TransNetR [11], and DilatedSegNet [12] are used for comparison. Additionally, performance comparisons with polyps generated in the existing polyp generation model [20] are also conducted.
The remainder of this paper is organized as follows. In Sect. 2, the proposed generation model, the segmentation model used in the experiment, and the experimental data are introduced. In Sect. 3, the quality of images generated by the generative model is discussed. Experimental results of the segmentation model are presented. Finally, we conclude this study in Sect. 4.
2 Methods
2.1 SemanticPolypGAN
Figure 1 shows the concept of the image and mask generation of SemanticPolypGAN. The existing SemanticStyleGAN uses fixed elements such as eyes, nose, and mouth in face images.
Proposed SemanticStyleGAN-based polyp image and mask generation framework. The polyp and mask image pass through the multilayer perceptron (MLP) and is mapped into the W space. Each W code is used to modulate the weight of the local generator. The local generator outputs a feature map and a pseudo-depth map. Each output pseudo-depth map is used to generate a mask for each class, and then these are combined to generate the overall mask m. The feature map and mask for each class are combined to generate the overall feature map f, which goes through RenderNet to generate a polyp image. Finally, the discriminator is trained using the generated images and masks
However, the position, size, and shape of the polyp and the non-polyp part are not fixed in the polyp image. Therefore, it is difficult to control the characteristics of polyps with the existing SemanticStyleGAN model. To solve this problem, we propose SemanticPolypGAN, which optimizes the model structure for polyp images. Using SemanticPolypGAN, the polyp mask and non-polyp mask are used as inputs. It can adjust the polyp, non-polyp, and background parts (black background part of the four corners of the polyp image). Figure 2 is an image used to train the proposed SemanticPolypGAN model. From the left, are the polyp, polyp mask, and non-polyp mask images. The non-polyp mask image is created by inverting the polyp mask image. The background part is generated automatically during training, excluding the polyp and non-polyp masks.
In Fig. 1, input images and mask images are entered into a multilayer perceptron (MLP) to map randomly sampled codes into W space [25]. The W code is used to modulate the weight of the local generator. \(W_{background}\) is the remaining portion excluding the polyp mask and non-polyp mask. \(W_{polyp}\) is the polyp portion, and \(W_{Non-Polyp}\) is the non-polyp portion, that is, the colon surface without polyps.
Local generators \(g_{background}\), \(g_{polypand}\), \(g_{non-polyp}\) of the background, polyp, and non-polyp parts are generated to control the shape and texture of each. Each local generator outputs feature maps \(f_{background}\), \(f_{polyp}\), \(f_{non-polyp}\), and pseudo-depth maps \(d_{background}\), \(d_{polyp}\), \(d_{non-polyp}\). Here, the pseudo-depth map has a similar structure to the z-buffering rather than an exact depth map. The z-buffering process stores depth information to determine the pixel that must be drawn higher when different objects are drawn at the same pixel. In this study, the polyp must be placed on the top of the non-polyp.
In the existing SemanticStyleGAN, the background shape is fixed using face image data, thus there is no need to output and train a pseudo-depth map from \(g_{background}\). Because this study uses polyp data with a variety of backgrounds, the background is also trained by outputting a pseudo-depth map from \(g_{background}\). Using the output pseudo-depth maps, masks \(m_{background}\), \(m_{polyp}\) and \(m_{non-polyp}\) for each class are generated, and these are combined to generate the overall mask m. Thereafter, the entire feature map f is generated through the Hadamard product of the feature map and masks for each class. RenderNet refines the entire mask m into a high-resolution segmentation mask and generates a polyp image. Finally, a discriminator is trained using the generated images and masks.
2.2 Network Architectures
Figure 3 is the architecture of the local generator used in SemanticPolypGAN. In SemanticStyleGAN, a coarse structure is placed in the local generator and used to control the overall part of the image. However, the coarse structure is unnecessary in polyp images because the position, size, and shape of the normal parts and polyps are not fixed. Therefore, the number of training parameters is reduced by removing coarse layers. To improve the quality of the generated polyps, the number of structure and texture layers is increased from four to six. Each layer is a 1\(\times\)1 convolution layer. The shape and texture latent codes are contained in \(w_s^k\) and \(w_t^k\), respectively. Here, w means W space, and k represents the polyp and non-polyp background, s represents shape, and t represents texture.
We use the Fourier feature map [26] for position encoding, to better train features by emphasizing the high-frequency components of the input data. First the shape and texture latent codes \(w_s^k\), \(w_t^k\), and \(p\) are input to the local generator \(g_k\). Thereafter, the structure layer passes through the toDepth layer, a linear fully connected layer, and outputs a 1-channel pseudo-depth map \(d_k\). Finally, the texture layer passes through the toFeat layer, a linear fully connected layer, and outputs a feature map \(f_k\) with 512 channels. Using Eq. 1, \(d_k\) and \(f_k\) can be calculated.
Figure 4 is the RenderNet structure proposed by SemanticPolypGAN. The output of RenderNet is adjusted according to the input feature map. It is very similar to the generator of StyleGAN2 [25] in that it uses a ConvBlock composed of two convolution layers. In this study, to better generate the features of small polyps, upsampling is started at 8 \(\times\) 8 by reducing the input feature map size from the existing 16 \(\times\) 16. The feature map is also concatenated on all blocks during upsampling. During upsampling, the entire mask image is also refined into a high-quality image.
The proposed local generator and RenderNet structure achieved better FID (Frechet inception distance) and IS (inception score) than the existing model when generating polyp images. (Refer to Sect. 3.2)
2.3 Polyp-Segmentation Model
To compare model performance based on the generated images, we used the latest polyp-segmentation models, UACANet [8], PraNet [9], TGANet [10], TransNetR [11], and DilatedSegNet [12]. We used polyp images generated by SemanticPolypGAN as additional training data for the five segmentation models to compare and evaluate the performance
2.4 Experimental Datasets
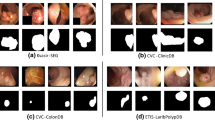
We used two sets of data for training of SemanticPolypGAN. One is 560 of the 612 images of the CVC-ClinicDB [14] dataset used in the Medical Image Computing and Computer-Assisted Intervention 2015 Colonoscopy Automatic Polyp Detection Challenge. The other is 880 of the 1000 images of the Kvasir-SEG [15] dataset released by Simula for research and education purposes. The remaining samples from each dataset were used for testing. To augment the data, shearing, translation, and 80% zoom were first applied, followed by 90-degree, 180-degree, and 270-degree rotation, up-down, left-right, and left-right symmetry, and finally, transpose augmentation. The same augmentation was applied to the mask images.
For training segmentation models, we used 1000 sheets of BKAI-IGH NeoPolyp-Small [16] and 880 sheets of Kvasir-SEG [15], which are publicly available. We generated 350 polyp images using SemanticStyleGAN and added them to the training data. For comparison, 350 polyp images generated in [20] were also used as additional training data to train each polyp-segmentation model. For testing, 52 images from CVC-ClinicDB [14], 120 images from Kvasir-SEG [15], and 300 images from CVC-300 [17] were used.
3 Results and Discussion
3.1 Generated Polyp Images and Masks
Figure 5 shows polyp and mask images generated by SemanticPolypGAN. The yellow part of the mask is the non-polyp, and the blue part is a polyp. The color, shape, and texture of the generated polyps are diverse and naturally match with the non-polyp parts. The generated background is diverse due to augmentations applied to the training image. There is white text at the top left, bottom left, and center of the image because there are many images with white text in the Kvasir-SEG data among the training images.
3.2 Generation Quality Evaluation
Table 1 shows the comparison of polyp-image quality generated after training with SemanticStyleGAN and SemanticPolypGAN. We used FID [27] and IS [28] as performance indicators. FID compares the quality and diversity of image sets by measuring the statistical distance between generated images and real images. IS evaluates model performance by predicting generated images by class through the inception network and using the entropy of the group. The first model was trained by inputting polyp images and masks into the SemanticStyleGAN. The second was trained by applying only RenderNet modification to SemanticStyleGAN. The final structure was trained using SemanticPolypGAN.
Results showed that when only RenderNet was modified, the performance was second best with FID and average IS of 21.77 and 3.81, respectively. When trained using the proposed SemanticPolypGAN, the performance was the best with FID and average IS of 20.64 and 3.91, respectively.
3.3 Shape and Texture Control of Polyp Images Through Latent Interpolation
SemanticPolypGAN can change the shape and texture of a specific semantic area by changing the latent code. Fig. 6 is the result of interpolating the background, polyp, and non-polyp areas of the image generated by SemanticPolypGAN. The polyp image and mask in 1st and 2nd rows are generated images to which interpolation is applied. Unlike SemanticStyleGAN, SemanticPolypGAN allows background interpolation. The background part of a colonoscopy image may vary depending on the endoscope camera or shooting environment. Thus, it can be transformed into an appropriate environment through interpolation or semantic synthesis. The black border background of the 1st row changes while the background texture does not change because it is all black.
Random latent interpolation results. The 1st and 2nd columns show the generated polyp images and masks. The 3rd and 5th row show transformed images after applying a random latent interpolation to the 1st row image, and the 4th and 6th row show transformed mask images. The 1st row shows the shape of the background. The 2nd row shows the shape of the non-polyp. The 3rd row shows the texture of the non-polyp part. The 4th row shows the shape of the polyp. The 5th row shows the result of randomly transforming the latent of the polyp texture
In the 2nd row, the shape part of the non-polyp shows slight changes in the size of surface wrinkles and holes. The non-polyp part in 3rd row is changed to various textures for the same polyp. In the 4th row, the shape of the polyp varies from a large polyp to a very small polyp. In the 5th row, the texture can be adjusted for a polyp of the same shape.
3.4 Semantic Synthesis Between Generated Polyp Images
Figure 7 below shows the result of the semantic synthesis between the generated polyp images. Images in 1st row, 1st column, and 2nd column were generated by SemanticPolypGAN. In the 1st column is the target image to which semantic synthesis was applied, and in the 1st row is the image used for semantic synthesis. SemanticPolypGAN can control the basic background, non-polyp, and polyp respectively. The 3rd and 4th columns show the results of compositing the background; the 5th and 6th columns show the results of compositing the non-polyp part, while the 7th and 8th columns show the results of compositing only the polyp part. The shape of the polyp in 7th column and 2nd row has enlarged, and the color of the polyp has also changed.
Result of semantic synthesis of the shape and texture of the background, non-polyp, and polyp at the same time. The images in the 1st row, 1st column, and 2nd column were generated by SemanticPolypGAN. The image in the 1st column is the image to which semantic synthesis was applied, and the image in the 1st row is the image used for semantic synthesis
SemanticPolypGAN can also control the shape and texture characteristics of each element. Figure 8 shows the results of compositing the shape and texture of non-polyp and polyp parts, respectively. The images in 1st row, 1st column, and 2nd column of Fig. 8 were generated by SemanticPolypGAN. In 2nd row and 5th column of (a), the texture of the polyp changed to show bleeding like the polyp in 1st row used for synthesis. Rather than simply using images generated by SemanticPolypGAN, polyps with more diverse features can be generated by semantic synthesis between images.
a Result of semantic synthesis of the polyp part, and b result of semantic synthesis of the shape and texture of the non-polyp part. The images in the 1st row, 1st column, and 2nd column were generated by SemanticPolypGAN. In the 1st column is the target image to which semantic synthesis was applied, and the image in the 1st row is the image used for semantic synthesis
3.5 Evaluation of Segmentation
Tables 2 and 3 show the results of training the five polyp-segmentation models using only the original training images (Original) and adding 350 images generated in [20] and 350 images generated by SemanticPolypGAN to the original images. Evaluation indicators of intersection-over-union (IoU) and Dice were used. Table 2 shows the results of comparing the performance of CVC-300, CVC-ClinicDB, and Kvasir-SEG as test sets after training using generated polyp images combined with BKAI-IGH data as the original training set.
Adding images generated using the proposed method to the training set improved performance compared to using only the original training set for all models. When the TransNetR model was tested on CVC-300 data, mean Dice showed the greatest performance improvement with a difference of 0.1003 compared to the original data. When 350 polyp images generated by the proposed method and the existing method [20] were added to training data, the mean IoU and mean Dice performance of the proposed method improved in 14 out of 15 experiments.
Table 3 shows the performance results of CVC-300, CVC-ClinicDB, and Kvasir-SEG test sets trained using generated images combined with Kvasir-SEG dataset. In 14 of 15 experiments (excluding the CVC-ClinicDB dataset test in the TransNetR model), the performance improved compared with that using only the original training set. When the DilatedSegNet model was tested on the CVC-300 data, the mean IoU and mean Dice showed the greatest improvement with differences of 0.0641 and 0.0609, compared with using the original set. When 350 polyp images generated by the proposed method were added in 14 experiments, the mean IoU and mean Dice were better than those of the existing method [20].
Figure 9 shows two examples: failure to segment a test image when trained with the original training set, and a successful segmentation after adding 350 images generated by SemanticPolypGAN. (a) is the original image of the Kvasir-SEG test set, and (b) is the corresponding polyp ground truth mask of (a). The mask result after training the PraNet model with the BKAI-IGH original training set and testing the image in (a) is shown in (c). The mask result of testing the image in (a) is shown in (d) after training the PraNet model by combining the BKAI-IGH original training set and 350 images generated by SemanticPolypGAN. A significant difference from the ground truth mask is shown in mask image (c); however, the mask image shows similar results to the ground truth mask in (d). This shows that the images generated by SemanticPolypGAN improve model performance.
3.6 Limitations and Future Work
Many polyp images can be generated using the proposed model, using semantic synthesis between the generated polyp images, a variety of polyp images can be generated. Fig. 10 shows performance improvement when generated polyp images are additionally added to the training set. For the UACANet and TGANet models, which showed good results in the previous polyp-segmentation performance evaluation in Tables 2 and 3, mIoU improved when the generated polyp images added to the original Kvasir-SEG data were increased by 200 to 600. The experiments confirmed that adding generated images improved the performance of both models. However, segmentation performance does not continue to improve with the addition of more generated images to training set. The performance of TGANet improved significantly when the number of generated images increased from 200 to 400; however, adding 600 images slightly improved the performance. The performance of UACANet improved the most with the addition of 200 images; after adding 400 images, there was no further improvement. Rather a slight decrease was observed. We believe that performance improvement varies with the number of images generated due to differences in the model size e.g., the number of training parameters for each model.
Change in performance due to the addition of images generated using the method proposed in this paper is shown in a and b. a mIoU change when training the UACANet model by adding 200, 400, and 600 generated images to Kvasir-SEG training set and b change in mIoU when the TGANet model is trained by adding 200, 400, and 600 generated images to Kvasir-SEG training set
Figure 11 shows two poorly segmented images from the results of training the UACANet model by adding 350 generated polyp images to the Kvasir-SEG data and testing them on CVC-300 data. The original image of CVC-300 test data is shown in (a), the ground truth polyp mask is shown in (b), and the prediction mask is shown in (c). In (c), the location is found to some extent, however, the division is not accurate. Thus, it is still difficult to segment polyp images with small shapes or unclear features. This might be caused by not having many such images in the training set and generated images.
4 Conclusion
It is difficult and expensive to collect sufficient training data and labels for deep-learning-based colonoscopy polyp-image segmentation. Therefore, we propose SemanticPolypGAN to generate colonoscopy polyp images. In existing polyp-generation models, input condition preparation steps are required, and it is difficult to independently control semantic elements during generation. SemanticPolypGAN uses only polyp images and masks as input images and controls the shape and texture of polyps and non-polyp parts when generating images. We compared the segmentation performance of five models between training on original data and training by adding generated images. Adding generated images improved polyp-segmentation performance for all models. The proposed model outperformed existing polyp-generation models in polyp segmentation.
Data Availability
This research utilizes four publicly available datasets and details are given in Sect. 2.4.
References
Colorectal Cancer. (2020). Technical report. International Agency for Research on Cancer. https://www.iarc.who.int/cancer-type/colorectal-cancer/
Cancer Stat Facts: Colorectal Cancer. (2023). Technical report. National Cancer Institute. https://seer.cancer.gov/statfacts/html/colorect.html
Zauber, A. G., Winawer, S. J., O’Brien, M. J., Lansdorp-Vogelaar, I., Ballegooijen, M., Hankey, B. F., Shi, W., Bond, J. H., Schapiro, M., Panish, J. F., & Stewart, E. T. (2012). Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. New England Journal of Medicine, 366(8), 687–696. https://doi.org/10.1056/NEJMoa1100370
Ahn, S. B., Han, D. S., Bae, J. H., Byun, T. J., Kim, J. P., & Eun, C. S. (2012). The miss rate for colorectal adenoma determined by quality-adjusted, back-to-back colonoscopies. Gut and Liver, 6(1), 64. https://doi.org/10.5009/gnl.2012.6.1.64
Urban, G., Tripathi, P., Alkayali, T., Mittal, M., Jalali, F., Karnes, W., & Baldi, P. (2018). Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology, 155(4), 1069–1078. https://doi.org/10.1053/j.gastro.2018.06.037
Shin, Y., Qadir, H. A., Aabakken, L., Bergsland, J., & Balasingham, I. (2018). Automatic colon polyp detection using region based deep cnn and post learning approaches. IEEE Access, 6, 40950–40962. https://doi.org/10.1109/ACCESS.2018.2856402
Wang, P., Xiao, X., Glissen Brown, J. R., Berzin, T. M., Tu, M., Xiong, F., Hu, X., Liu, P., Song, Y., Zhang, D., & Yang, X. (2018). Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nature Biomedical Engineering, 2(10), 741–748. https://doi.org/10.1038/s41551-018-0301-3
Kim, T., Lee, H., & Kim, D. (2021). Uacanet: Uncertainty augmented context attention for polyp segmentation. In Proceedings of the 29th ACM international conference on multimedia (pp. 2167-2175). https://doi.org/10.1145/3474085.3475375
Fan, D. P., Ji, G. P., Zhou, T., Chen, G., Fu, H., Shen, J., & Shao, L. (2020). Pranet: Parallel reverse attention network for polyp segmentation. In International conference on medical image computing and computer-assisted intervention (pp. 263-273). Springer. https://doi.org/10.1007/978-3-030-59725-2_26
Tomar, N. K., Jha, D., Bagci, U., & Ali, S. (2022) TGANet: Text-guided attention for improved polyp segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 151-160). Springer. https://doi.org/10.1007/978-3-031-16437-8_15
Jha, D., Tomar, N. K., Sharma, V., & Bagci, U. (2023). TransNetR: Transformer-based residual network for polyp segmentation with multi-center out-of-distribution testing. Preprint retrieved from https://arxiv.org/abs/2303.07428
Tomar, N. K., Jha, D., & Bagci, U. (2023). Dilatedsegnet: A deep dilated segmentation network for polyp segmentation. In International conference on multimedia modeling (pp. 334-344). Springer. https://doi.org/10.1007/978-3-031-27077-2_26
Abouelmehdi, K., Beni-Hssane, A., Khaloufi, H., & Saadi, M. (2017). Big data security and privacy in healthcare: A review. Procedia Computer Science, 113, 73–80. https://doi.org/10.1016/j.procs.2017.08.292
Bernal, J., Sánchez, F. J., Fernández-Esparrach, G., Gil, D., Rodríguez, C., & Vilariño, F. (2015). Wm-dova maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Computerized Medical Imaging and Graphics, 43, 99–111. https://doi.org/10.1016/j.compmedimag.2015.02.007
Jha, D., Smedsrud, P. H., Riegler, M. A., Halvorsen, P., De Lange, T., Johansen, D., & Johansen, H. D. (2020). Kvasir-seg: A segmented polyp dataset. In MultiMedia modeling: 26th international conference, MMM 2020, Daejeon, South Korea, January 5–8, 2020, proceedings, part II 26 (pp. 451-462). Springer. https://doi.org/10.1007/978-3-030-37734-2_37
Ngoc Lan, P., An, N. S., Hang, D. V., Long, D. V., Trung, T. Q., Thuy, N. T., & Sang, D. V. (2021). Neounet: Towards accurate colon polyp segmentation and neoplasm detection. In Advances in visual computing: 16th international symposium, ISVC 2021, virtual event, October 4-6, 2021, proceedings, part II (pp. 15-28). Springer. https://doi.org/10.1007/978-3-030-90436-4_2
Vázquez, D., Bernal, J., Sánchez, F. J., Fernández-Esparrach, G., López, A. M., Romero, A., Drozdzal, M., & Courville, A. (2017). A benchmark for endoluminal scene segmentation of colonoscopy images. Journal of Healthcare Engineering. https://doi.org/10.1155/2017/4037190
Adjei, P. E., Lonseko, Z. M., Du, W., Zhang, H., & Rao, N. (2022). Examining the effect of synthetic data augmentation in polyp detection and segmentation. International Journal of Computer Assisted Radiology and Surgery, 17(7), 1289–1302. https://doi.org/10.1007/s11548-022-02651-x
Shin, Y., Qadir, H. A., & Balasingham, I. (2018). Abnormal colon polyp image synthesis using conditional adversarial networks for improved detection performance. IEEE Access, 6, 56007–56017. https://doi.org/10.1109/ACCESS.2018.2872717
Qadir, H. A., Balasingham, I., & Shin, Y. (2022). Simple u-net based synthetic polyp image generation: Polyp to negative and negative to polyp. Biomedical Signal Processing and Control, 74, 103491. https://doi.org/10.1016/j.bspc.2022.103491
Isola, P., Zhu, J. Y., Zhou, T., & Efros, A. A. (2017). Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 1125-1134). https://arxiv.org/abs/1611.07004
Mirza, M., & Osindero, S. (2014). Conditional generative adversarial nets. https://arxiv.org/abs/1411.1784
Karras, T., Laine, S., & Aila, T. (2019). A style-based generator architecture for generative adversarial networks. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 4401-4410).
Shi, Y., Yang, X., Wan, Y., & Shen, X. (2022). Semanticstylegan: Learning compositional generative priors for controllable image synthesis and editing. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 11254-11264). https://arxiv.org/abs/2112.02236
Karras, T., Laine, S., Aittala, M., Hellsten, J., Lehtinen, J., & Aila, T. (2020). Analyzing and improving the image quality of stylegan. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 8110-8119). https://arxiv.org/abs/1912.04958
Tancik, M., Srinivasan, P., Mildenhall, B., Fridovich-Keil, S., Raghavan, N., Singhal, U., Ramamoorthi, R., Barron, J., & Ng, R. (2020). Fourier features let networks learn high frequency functions in low dimensional domains. Advances in Neural Information Processing Systems, 33, 7537–7547.
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B., & Hochreiter, S. (2017). Gans trained by a two time-scale update rule converge to a local nash equilibrium. Advances in Neural Information Processing Systems, 30, 1–12.
Salimans, T., Goodfellow, I., Zaremba, W., Cheung, V., Radford, A., & Chen, X. (2016). Improved techniques for training gans. Advances in Neural Information Processing Systems, 29, 1–9.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Education(2022R1I1A3063458)
Author information
Authors and Affiliations
Contributions
YHS conceived the idea and verified the analytical methods. The experiments and manuscript preparation were conducted by HS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
Not applicable.
Informed Consent
Not applicable.
Consent for Publications
Not applicable.
Research Involving in Human and Animal Participants
This study does not include human or animal participants.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, H., Shin, Y. Semantic Polyp Generation for Improving Polyp Segmentation Performance. J. Med. Biol. Eng. 44, 280–292 (2024). https://doi.org/10.1007/s40846-024-00854-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-024-00854-y