Abstract
Purpose
To evaluate the classification performance of structured report features, radiomics, and machine learning (ML) models to differentiate between Coronavirus Disease 2019 (COVID-19) and other types of pneumonia using chest computed tomography (CT) scans.
Methods
Sixty-four COVID-19 subjects and 64 subjects with non-COVID-19 pneumonia were selected. The data was split into two independent cohorts: one for the structured report, radiomic feature selection and model building (n = 73), and another for model validation (n = 55). Physicians performed readings with and without machine learning support. The model's sensitivity and specificity were calculated, and inter-rater reliability was assessed using Cohen’s Kappa agreement coefficient.
Results
Physicians performed with mean sensitivity and specificity of 83.4 and 64.3%, respectively. When assisted with machine learning, the mean sensitivity and specificity increased to 87.1 and 91.1%, respectively. In addition, machine learning improved the inter-rater reliability from moderate to substantial.
Conclusion
Integrating structured reports and radiomics promises assisted classification of COVID-19 in CT chest scans.
Similar content being viewed by others
1 Introduction
Computed tomography is a significant tool in the diagnosis and staging of pneumonia, providing important information for the management of the recent COVID-19 pandemic [1]. However, aspects relating to pulmonary lesions seen in CT are typical but not pathognomonic [2], suggesting that this method has its limitations [3].
Artificial intelligence (AI) is commonplace in medical imaging due to its ability to automate image segmentation, improve lesion detection, better perform quality assurance, and increase inter- and intra-rater reliability [4,5,6,7]. Moreover, structured reports are claimed to have enhanced radiology reporting by enabling standardization, productivity, and improved information transmission [8, 9], leveraging their potential to support decision-making technologies using AI [8, 10, 11].
Recent studies have shown promising trends for radiomics and AI in classifying pneumonia in CT studies [12, 13]. Even though AI has been widely used with different data types for COVID-19 diagnoses, such as radiomics and clinical data, it has been little used in respect of imaging reports [14]. Huanhuan Liu et al. developed a clinical-radiomics model that included distribution features and clinical data, demonstrating the added value of lesion distribution reporting in their models [13]. In this scenario, the integration of minable data with artificial intelligence can provide tangible benefits for radiology practices and patient care.
In this work, we explored radiological features from multi-center, standardized web-based structured reports and radiomics. We also developed AI models and evaluated their classification performance to differentiate COVID-19 from other types of pneumonia, while evaluating their impact on intra-rater reliability.
2 Materials and Methods
2.1 Subjects
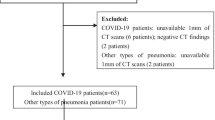
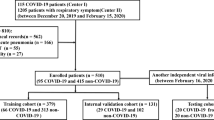
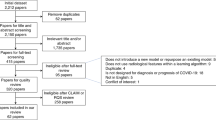
Cases were defined as subjects with COVID-19 confirmed using RT-PCR and referred for thorax CT between April, 2020 and April, 2021 and sequentially selected from three Brazilian academic hospitals (Table 1). Controls were defined as subjects sequentially selected with pneumonia before the COVID-19 pandemic, having been referred for thorax CT between January, 2018 and October, 2019.
Exclusion criteria were (a) CT images with poor quality; (b) small or imperceptible lesions; (c) unavailability of RT-PCT for the confirmed COVID-19 group; (d) images that were not compliant with the institutional protocol for lung imaging (e.g., matrix size, kernel); and (e) images that were no longer available on the Picture Archiving Communication System (PACS) during the retrieval process.
The study was approved by the ethics committee at each institution: Hospital Universitario Prof. Edgard Santos (UFBA) (4.494.511), Hospital Universitario Alcides Carneiro (UFCG) (4.569.389), and Hospital Universitario de Juiz de Fora (UFJF) (4.926.688).
2.2 Structured Report
The structured report was composed of ten questions concerning the radiological findings: five concerning lesion analysis and five concerning lesion distribution, eliciting the following binary (yes/no) responses:
Analysis:
-
A1: Predominant ground-glass opacity with a rounded morphology.
-
A2: Ground-glass opacities with superimposed interlobular septal thickening and intralobular septal thickening (crazy-paving pattern).
-
A3: Ground-glass opacity and pulmonary consolidation.
-
A4: Pulmonary consolidation with air bronchograms.
-
A5: Reversed halo sign or cryptogenic organizing pneumonia (COP).
Distribution:
-
D1: Bilateral and multifocal.
-
D2: Peripheral distribution.
-
D3: Prevalent in lower lobes and dorsal region.
-
D4: Peribronchovascular opacities and peripheral distribution.
-
D5: Diffuse opacities.
Experienced radiologists (one from each hospital) retrieved the corresponding CT images from the hospital PACS and selected the corresponding structured report findings. Only image series with axial slices of the lung were considered. This data was then included in the Nuclearis software (Salvador, Brazil), a web-based radiology information system capable of personalizing standard structured reports. No other information, such as side notes, remarks, additional details, was analyzed.
2.2.1 Structured Report Model
An Extreme Gradient Boosting (XgBoost) algorithm was built into the training cohort (n = 73) and validated in the independent cohort (n = 55) using a structured report score of 0 or 1, with 0 representing non-COVID-19 and 1 representing COVID-19.
2.3 Radiomics
2.3.1 Automatic Segmentation
The process involved 37 COVID-19 individuals and a further 36 individuals with pneumonia not associated with COVID-19 (hereafter denominated feature selection cohort). CT images of these patients had been manually segmented using LIFEx, version 6.30 (www.lifex.org), to train a 2D convolutional neural network [15] with a fivefold cross-validation scheme. A total of 12,780 CT slices were used, and data were augmented using 1 mm erosion in the LIFEx ROIs, as well as flip and rotation in the original images and ROIs, which resulted in a total of 51,120 images. Each training dataset used 60 subjects to segment the additional subjects automatically. The 2D output probability maps were filtered with a 3D Gaussian kernel before thresholding to obtain the final 3D lesion segmentation. Only the independently segmented lesions were included in the radiomic feature selection analysis. This process used the established image segmentation software, LIFEx, to delineate the ground truth of lung lesions with the aim of developing proprietary automatic segmentation software for the radiomic feature extraction.
2.3.2 Feature Extraction
Radiomic features were extracted using PyRadiomics [16]. The matrix grid was resampled to voxels of 1×1×2 mm3, and the discretization within each ROI was scaled to 128 grayscale levels. Radiomic feature classes and corresponding features are presented in the supplementary material. A total of 93 features were calculated, including 18 first order and 75 textural features: 16 Gy level run length matrix (GLRLM), 16 Gy level size zone matrix (GLSZM), five neighborhood gray-tone difference matrix (NGTDM), 14 Gy level dependence matrix (GLDM), and 24 Gy level co-occurrence matrix (GLCM).
2.3.3 Feature Selection
The feature selection process is illustrated in Fig. 1. Lung lesions from the feature selection cohort were automatically segmented, and two additional segmentation datasets (artificial observers) were generated using erosion functions of 1 and 3 mm. Additional blur and sharp kernels were applied to the original image to simulate different scanner/image reconstructions. Ninety-three radiomic features were computed for each of the three observers from the erosion and image quality groups. The intraclass correlation coefficient (ICC) was used to estimate reproducibility. Features which ICC > 0.90 were selected.
2.3.4 Radiomic Model
We used the same cohort of feature selection for model building (n = 73) and an independent cohort with 55 subjects (27 COVID-19 and 28 non-COVID-19) for model validation. A random forest model was built at the lesion level (segmented lesions = 1060), and annotated according to subject status. The algorithm classified lesions using a radiomic score that ranged from 0 to 1, with 0 representing non-COVID-19 and 1 representing COVID-19 lesion. The model (rf mean) was validated at the subject level by averaging the scores of all lesions for the specific subject.
2.4 Physician’s Performance
Two experienced physicians, one radiologist, and one pneumologist scored the CT images in the validation group (n = 55) to classify the pneumonia as COVID-19 or non-COVID-19. Both physicians were blinded in respect of RT-PCR and clinical information. The radiologist was the same person that performed the structured report (Hospital B), and the pneumologist only participated in this validation step. They initially analyzed the CT images independently, with no support from the machine learning models. After 30 days they repeated the analysis with the support of both radiomic and structured report models using a binary classification for each model.
2.5 Statistical Analysis
The odds ratios of radiological findings were assessed as an indication of feature importance, using the entire dataset (n = 128). The mean of Dice Coefficient (F1 Score) was computed using Python 3.9.2 software to assess the performance of our automatic COVID-19 segmentation method. Machine learning analysis was performed using Orange software, version 3.29.3 [17]. The sensitivity and specificity of the models were calculated according to category classification outputs from Orange, and the 95% confidence intervals were estimated using OpenEpi software [18]. The receiver operating characteristics (ROC) curves were assessed to estimate the area under the ROC curve (AUC) and the 95% confidence intervals (CI) using the continuous scores from Orange and SPSS 18.0 for Windows. The mean ROC curve for physicians’ performance was simulated by iteratively misspecifying the true values with a random gaussian function to enable improved readability and interpretation. The procedure was stopped when the simulated ROC curve yielded values corresponding to the real sensitivity and specificity [19]. The inter-rater reliability was assessed with and without the support of machine learning models using Cohen’s Kappa coefficient (k), with the following classification: weak (k ≤ 0.20), fair (0.20 < k ≤ 0.40), moderate (0.40 < k ≤ 0.60), substantial (0.60 < k ≤ 0.80) or strong (k > 0.80) [20].
3 Results
3.1 Structured Reports
The XgBoost performed with 81.6% (66.6–90.8%) sensitivity and 82.9% (67.3–91.9%) specificity in the training cohort (AUC = 0.91) and 77.3% (56.6–89.9%) sensitivity and 69.7% (52.7–82.7%) specificity in the validation cohort (AUC = 0.82) (Table 3).
The odds ratio (95% CI) revealed feature importance, as presented in Table 2. According to this analysis, features A1, A2, A4, D1, and D2 were the strongest predictors (p < 0.05), being present with greater frequency in the COVID-19 group, except for A4, which appeared more frequently in the non-COVID-19 group.
3.2 Radiomics
The mean F1 score of the auto-segmentation algorithm was 0.72. Figure 2 shows the axial view of an infected COVID-19 individual (Fig. 2a), the auto-segmentation (Fig. 2b), ROI erosion functions at 1 mm (Fig. 2c) and 3 mm (Fig. 2d), as well as image enhancements of blurred (Fig. 2e) and sharp (Fig. 2f) kernels.
The intersection between erosion and image quality groups resulted in three features showing excellent reproducibility: GLDM Gray Level Non-Uniformity, First Order Energy, and First Order Total Energy.
The rf mean model with these selected features produced AUC = 1.0 in the training cohort and AUC = 0.84 (0.73–0.95) in the validation cohort, both at the subject level. Sensitivity and specificity in the validation cohort were 70.4% (51.5–84.2%) and 89.3 (72.3–96.3%), respectively (Table 3).
3.3 Physicians
In respect of the physicians’ assessment, the pneumologist performed with sensitivity and specificity of 81.5% (63.3–91.8%) and 67.9% (49.3–82.1%), respectively; while the radiologist performed with sensitivity and specificity of 85.2% (67.5–94.1%) and 60.7% (42.4–76.43%), respectively. When physicians were assisted with artificial intelligence models, their performance for sensitivity and specificity were, respectively, 85.2% (67.5–94.1%) and 85.7 (68.5–94.3) for the pneumologist, and 88.9% (71.9–96.2%) and 96.4% (82.3–99.4%) for the radiologist. The Cohen’s Kappa coefficient for both physicians was k = 0.66 (0.46–0.86) without the support of machine learning, which rose to 0.78 (0.61–0.94) with the use of machine learning. Therefore, the readings that involved assistance by artificial intelligence improved the lower limit, which increased from moderate to substantial. Figure 3 depicts the overall readers’ performance based on the AUC, while Fig. 4 depicts the readers’ reliability.
Even though we only found a significant difference in specificities for the radiologist reader with and without AI (p < 0.05), our results suggest that standardized and semi-automatic measures based on questionnaires and radiomics may assist physicians in respect of classification.
4 Discussion
This study shows that structured reports, radiomics, and machine learning algorithms provide a reliable classification of CT scans when identifying COVID-19 individuals and better assist physicians in their decisions. Our study characterized CT pneumonia examinations using radiomics and a typically structured report questionnaire for lung CT pneumonia and AI models. Furthermore, we conducted a careful methodological and statistical design to avoid pitfalls in the modeling workflow and analysis, thus promoting the results' reliability and repeatability [21, 22].
Syeda et al. [14] published a systematic review on the role of machine learning in tackling coronavirus disease. However, of the forty studies that have evaluated the diagnosis of COVID-19, none used imaging features extracted from standardized structured reports. Although several authors reported the application of AI for diagnoses of COVID-19 using CT, their works were based on image processing techniques and/or clinical data.
We were not able to demonstrate that the model’s performance was statistically higher than that achieved by the physicians, but the observed trends provide cause for optimism. More importantly, the physicians improved their reporting when assisted by these models. Interestingly, the results from our radiomic model are similar to Guiot J et al. [12], who achieved a sensitivity of 69.5% and a specificity of 91.6%. Liu H et al. [13] developed both clinical-radiological and clinical-radiomic models, reporting sensitivity and specificity of 63 and 84%, respectively, for their clinical-radiological model, which included the features of age, gender, neutrophil ratio, lymphocyte count, location, distribution, reticulation, and lung involvement. The overall performance of AUC = 0.83 (0.75–0.90) is also equivalent to our structured report model, while their clinical-radiomic model achieved AUC = 0.93 (0.85–1.00) with a sensitivity and specificity of 85 and 90%. Although clinical data may provide additional predictive information, such data are not available in many clinical settings, while our models require only simple scoring and automated image quantification.
Our work also confirmed the higher prevalence of typical features previously reported in COVID-19 cases (A1, A2, D1, D2) [1, 23], demonstrating the added contribution of these patterns and their dependence on artificial intelligence models. The performance of these models motivates a more extensive clinical evaluation based on a larger cohort designed to evaluate gains in accuracy and inter/intra-rater reliability by radiologists with different levels of experience using our algorithm.
Our study has certain limitations. Firstly, the limited sample size imposes a relatively large confidence interval for our results. Sensitivity and specificity ratios are not affected by the sample size, but the confidence interval in larger samples is smaller. Our results are presented with 95% confidence interval and 98% statistical power [18]. Secondly, our radiomic model could be generalized only using the feature selection and extraction parameters used in this work. In addition, our algorithm shows two scores (radiomic and structured reports) therefore limiting the capacity of the decisions taken by physicians when both scores are ambiguous (n = 18/55). Finally, our models are disease specific. The AI models were trained/validated using subjects affected by pneumonia, meaning that our results are not necessarily valid for cancer or other inflammatory lesions. Our method only informs the likelihood of the lesions’ etiology being due to COVID-19.
The management of COVID-19 patients is multifaceted. The CT aspects of COVID-19 lesions are usually presented with ground-glass opacity, rounded or polygonal, and its distribution is mostly peripheral and bilateral, without pulmonary consolidation (Table 2). Since inflammatory lesions in the lungs are not pathognomonic, the etiology of some pneumonia cases is difficult to differentiate using simply CT without complementary technologies. Thus, decision-making technologies to handle coronavirus disease in real-time are valuable tools to both avoid its transmission and also rationalize resources. The proposed approach intends to improve standardization, automate and identify infected patients as well as discard cases of pneumonia that are unlikely to involve COVID-19. Our efforts are aligned with the global demand to make minable data and artificial intelligence feasible and translational to clinical practice, given that radiomics, reporting standardization, and statistical tools are increasingly deployed [24].
5 Conclusion
Radiomics, structured reports, and machine learning algorithms enable good classification of CT scans to identify COVID-19 individuals. In addition, this approach assisted physicians in standardizing better the classification and reporting of COVID-19 in computed tomography.
Data availability
Information created during and/or analysed during the current study is available from the corresponding author upon reasonable request.
References
Kwee, T. C., & Kwee, R. M. (2020). Chest CT in COVID-19: What the radiologist needs to know. Radiographics, 40(7), 1848–1865.
da Rosa, M. R., Francelino Silva Junior, L. C., Santos Santana, F. M., Farias, T., & de Oliveira. (2021). Clinical manifestations of COVID-19 in the general population: systematic review. Wiener klinische Wochenschrift, 133(7–8), 377–82.
Barbosa, P. N. V. P., Bitencourt, A. G. V., Miranda, G. D., Almeida, M. F. A., & Rubens, C. (2020). Chest CT accuracy in the diagnosis of SARS-CoV-2 infection: Initial experience in a cancer center. Radiologia Brasileira, 53(4), 211–215.
Yasaka, K., & Abe. (2018). Deep learning and artificial intelligence in radiology: Current applications and future directions. PLoS medicine, 15(11), e1002707.
Dahl, F. A., Rama, T., Hurlen, P., Pål, H. B., Haldor, H., Tore, G., Øystein, N., & Lilja, Ø. (2021). Neural classification of Norwegian radiology reports: using NLP to detect findings in CT-scans of children. BMC Medical Informatics and Decision Making, 21(1), 1–8.
Yi, C., Tang, Y., Ouyang, R., Zhang, Y., Cao, Z., Yang, Z., Wu, S., Han, M., Xiao, J., Chang, P., & Ma, J. (2022). The added value of an artificial intelligence system in assisting radiologists on indeterminate BI-RADS 0 mammograms. European Radiology, 32(3), 1528–37.
Chan, K. S., Chan, Y. M., Tan, A. H. M., Shanying, L., Yuan, T. C., Qiantai, H., Enming, Y., Lester, R. C. C., Zhang, Li., Glenn, W. L. T., Sadhana, C., & Zhiwen, J. L. (2022). Clinical validation of an artificial intelligence-enabled wound imaging mobile application in diabetic foot ulcers. International Wound Journal, 19(1), 114–24.
Barbosa, F., Maciel, L. M. Z., Vieira, E. M., Paulo de Azevedo, M. M., Jorge, E., & Valdair, F. M. (2010). Radiological reports: a comparison between the transmission efficiency of information in free text and in structured reports. Clinics, 65(1), 15–21.
Reiner, B. I. (2009). The challenges, opportunities, and imperative of structured reporting in medical imaging. Journal of Digital Imaging, 22(6), 562–568.
Lakhani, P., Prater, A. B., Hutson, R. K., Andriole, K. P., Dreyer, K. J., Morey, J., Prevedello, L. M., Clark, T. J., Geis, J. R., Itri, J. N., & Hawkins, C. M. (2018). Machine learning in radiology: applications beyond image interpretation. Journal of the American College of Radiology, 5(2), 350–59.
Nguyen, D. H., & Patrick, J. D. (2014). Supervised machine learning and active learning in classification of radiology reports. Journal of the American Medical Informatics Association, 21(5), 893–901.
Guiot, J., Vaidyanathan, A., Deprez, L., Fadila, Z., Denis, D., Anne-Noëlle, F., Marie, T., Monique, H., Gregory, C., Stephane, M., Evanthia, E., Philippe, L., Nathan, T., Benjamin, M., Sean, W., Michel, M., Renaud, L., Paul, M., Wim, V., … Pierre, L. (2020). Development and validation of an automated radiomic CT signature for detecting COVID-19. Diagnostics, 11(1), 41.
Liu, H., Ren, H., Wu, Z., Xu, H., Shuhai, Z., Jinning, L., & Liang, H. (2021). CT radiomics facilitates more accurate diagnosis of COVID-19 pneumonia: compared with CO-RADS. Journal of Translational Medicine, 19(1), 1–12.
Syeda, H. B., Syed, M., Sexton, K. W., Shorabuddin, S., Salma, B., Farhanuddin, S., Fred, P., & Feliciano, Y. J. (2021). Role of machine learning techniques to tackle the COVID-19 crisis: Systematic review. JMIR Medical Informatics, 9(1), e23811.
Fan, D., Zhou, T., Ji, G., Zhou, Y., Chen, G., Fu, H., Shen, J., & Shao, L. (2020). Inf-net: Automatic COVID-19 lung infection segmentation from CT images. IEEE Transactions on Medical Imaging, 39(8), 2626–2637.
Van Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., Beets-Tan, R. G. H., Fillion-Robin, J. C., Pieper, S., Hugo, J. W. L., & Aerts. (2017). Computational radiomics system to decode the radiographic phenotype. Cancer Research, 77(21), e104-7.
Demsar, J., Curk, T., Erjavec, A., Gorup, Č, Hočevar, T., Milutinovič, M., Možina, M., Polajnar, M., Toplak, M., Starič, A., & Štajdohar, M. (2013). Orange: Data mining toolbox in python. Journal of Machine Learning Research., 14(35), 2349–53.
Dean AG, Sullivan KM, Soe MM. OpenEpi: Open Source Epidemiologic Statistics for Public Health [www.OpenEpi.com] [updated 2013 Apr 06; cited in 2021 Aug 21].
Kundu, S., Kers, J. G., & Janssens, A. C. J. (2016). Constructing hypothetical risk data from the area under the ROC curve: Modelling distributions of polygenic risk. PLoS ONE, 11(3), e0152359.
Sim, J., & Wright, C. C. (2005). The Kappa statistic in reliability studies: use, interpretation, and sample size requirements. Physical Therapy, 85(3), 257–268.
Vanwinckelen G and Blockeel H. On estimating model accuracy with repeated cross-validation. Benelearn 2012: Proceedings of the 21st Belgian-Dutch conference on machine learning. 2012: 39–44
Ying, X. (2019). An overview of overfitting and its solutions. J. Phys., 1168(2), 022022.
Adams, H. J., Kwee, T. C., Yakar, D., Hope, M. D., & Kwee, R. M. (2020). Chest CT imaging signature of coronavirus disease 2019 infection: In pursuit of the scientific evidence. Chest, 158(5), 1885–1895.
Buvat, I., & Orlhac, F. (2021). The TRUE checklist for identifying impactful artificial intelligence-based findings in nuclear medicine: is it true? Is it reproducible? Is it useful? Is it explainable? Journal of Nuclear Medicine, 62(6), 752–754.
Acknowledgements
This study was financed, in part, by Financiadora de Estudos e Pesquisas (Finep) (contract number 03.20.0156.00) for the financial support for Radtec Serviços em Física Médica to develop Covid-19 technological solutions; and by the Coordenacão de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001. The funder had no role in study design, data collection, data analysis, data interpretation, or writing of the report. We also appreciate Vinícius de Oliveira Menezes for his contribution with the computational infrastructure.
Author information
Authors and Affiliations
Contributions
MADM: contributed to study conception and design, data collection, data curation algorithm development, data analysis, interpretation of results, drafting the article, revising and final approval of the version to be published. RRES: contributed to study design, data curation, algorithm development, data analysis, revising and final approval of the version to be published. MN: contributed to study design, interpretation of results, revising and final approval of the version to be published. ASL, MCLCN, CTAS and ABLM: contributed to data analysis, interpretation of results, revising and final approval of the version to be published. DMO and TRR: contributed to data collection, data curation, revising and final approval of the version to be published. LMA and CZ: contributed to algorithm development, interpretation of results, revising and final approval of the version to be published. EMN: contributed to study conception and design, data analysis, interpretation of results, drafting the article, revising and final approval of the version to be published.
Corresponding author
Ethics declarations
Conflict of interest
Marcos A. D. Machado receives royalties from Radtec Serviços em Física Médica for artificial intelligence product innovations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Machado, M.A.D., Silva, R.R.E., Namias, M. et al. Multi-center Integrating Radiomics, Structured Reports, and Machine Learning Algorithms for Assisted Classification of COVID-19 in Lung Computed Tomography. J. Med. Biol. Eng. 43, 156–162 (2023). https://doi.org/10.1007/s40846-023-00781-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-023-00781-4