Abstract
Lumbar disc herniation is a common disease that causes low back pain. Due to the high cost of medical diagnosis, as well as a shortage and uneven distribution of medical resources, a system that can automatically analyze and diagnose lumbar spine Magnetic Resonance Imaging (MRI) is becoming an urgent need. This study uses deep learning methods to establish a classifier to diagnose lumbar disc herniation. An MRI classification dataset of lumbar disc herniation consisting of public MRI images is presented and is used to train the proposed classifier. Because a common difficulty in applying computer vision technology to medical images is labeling training data, we use a semi-supervised model training method, while multilayer transverse axial MRI images are used as the model input. In this method, we first use unlabelled MRI images for random self-supervised pre-training and the pre-trained model as a feature extractor for MRI images. Then, all marked cross-sections of each intervertebral disc are used to calculate the feature vector through the feature extractor. The information of all feature vectors is integrated, while a multilayer perceptron is used for classification training. After training, the model achieved 87.11\(\%\) accuracy, 87.50\(\%\) sensitivity, 86.72\(\%\) specificity and 0.9487 AUC (Area Under the ROC Curve) index on the test set. To analyze the rationality of the diagnostic results more quickly, we output the severity of degenerative changes in each region using a heatmap.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The intervertebral disc consists of a nucleus pulposus, a hydrous jelly-like substance, and an annulus fibrosus, which limits the position of the nucleus pulposus [1]. Typically, the annulus fibrosus confines the nucleus pulposus material within the ring apophysis of the vertebra. Spinal nerves travel outside the ring apophysis range, primarily in the central spinal canal, and diverge to the body through the intervertebral foramen on both sides of the spine. However, the intervertebral disc can break through the range of the ring apophysis due to trauma, prolonged external pressure, or dehydration caused by aging problems. Abnormal intervertebral discs can be divided into many categories according to their specific shapes, such as leakage of the nucleus pulposus material due to tearing of the annulus fibrosus, horizontal bulging of the intervertebral disc, and vertical protrusion of the Mohs nodules. The abnormal morphology of these discs may cause compression of the nerves in the central spinal canal or the nerve roots in the intervertebral foramen, which may lead to symptoms such as low back pain, leg pain, numbness, etc. [2]. Regardless of its specific shape, the similarity among these different abnormalities is that the intervertebral disc breaks through the ring apophysis and can compress the nerve. This situation is likely to be diagnosed as lumbar disc herniation [3].
The examination of lumbar intervertebral disc herniation by MRI generally requires an experienced radiologist to observe the shape of the intervertebral disc in the MRI image [4, 5], which is generally combined with sagittal images and transverse axial images concurrently. The transverse axis image primarily concerns the T2-weighted scanned image. For the automatic diagnosis of the lumbar disc herniation method, most existing studies only use the images of the central sagittal plane [6,7,8] or the paradigm of object detection to design the model [9, 10], labeling of intervertebral disc tissues of different morphologies and design of assisted diagnostic models using the paradigm of target detection and segmentation [11, 12], which may ignore much important information. However, the cross-sectional image contains more detailed information. The method of diagnosing lumbar disc herniation in the clinical guidelines is also defined from the perspective of the transverse axis [3]. By extracting multi-scale features from transaxial MRI, the spinal tissues can be segmented more accurately, and a higher recognition accuracy can be obtained [13, 14]. Therefore, to receive more practical information from the diagnostic model, we used the transverse axial lumbar spine MRI image to design the model. However, the images in the horizontal axis lack information in the vertical direction. Although the sagittal image can be introduced as it’s input, it may reduce the model’s ease of construction. Thus, we used another method by inputting multiple-layer images along the horizontal axis. If a single cross-section scanned image is used, the central cross-section of the intervertebral disc would be selected, and the MRI equipment will scan a cross-section with a given thickness, such as 4 mm. This cross-section will contain vertical information with a thickness of 4 mm. Also, if multiple layers of cross-sections are superimposed to form a three-dimensional scanned image, it can contain more vertical information. When the thickness of the three-dimensional image is sufficiently large, it can contain all sagittal images theoretically. In the actual design, we use the images of the three layers consisting of the central cross-section and its upper and lower adjacent layers as the model’s input, a method that several experiments have successfully verified.
In recent years, machine learning technologies represented by deep learning [15] have been increasingly applied in the field of computer-aided diagnosis (CAD) [16,17,18,19]. One of the most commonly used methods is supervised learning. When applied in medical imaging diagnosis, a common difficulty is acquiring numerous standardized labeled data. The data labeling process often requires the participation of radiologists in cooperation with algorithm researchers. Doctors may spend a large amount of time labeling data according to the specifications designed by algorithm researchers. The algorithm may undergo multiple revisions and iterations during the research process, and sometimes, the original data may be relabelled. Also, if the model needs good generalizability, we could ensure that the amount of the training data is sufficient; if insufficient, the model will be overfitted. Therefore, we use a training paradigm of semi-supervised learning [20] in this study. semi-supervised learning is a paradigm in which labeled and unlabelled data are used for model training to reduce the number of labeled samples while maintaining good generalizability. Semi-supervised learning need to choose different strategies according to a specific task. Compared with many realistic multimedia photos, MRI images of the lumbar spine have unique characteristics. The first is the significant similarity between images. All lumbar spine MRI images have similar structures, such as an intervertebral disc, a lamina, and surrounding adipose tissue. This type of “invariance” between images often does not constitute the basis for diagnosing diseases. However, the difference in details between images is the key information for the diagnosis, such as the level of the nucleus pulposus signal and the relationship between the intervertebral disc and the dural sac location. Considering this feature, we use a two-stage semi-supervised learning paradigm [21,22,23] based on contrastive self-supervised methods. According to the design purpose of the task, only the central cross-sectional scanned image of a portion of the patient’s intervertebral disc and its adjacent cross-sectional images will be labeled and used for classification. There are also many unlabelled MRI images in the training data. We thus conduct semi-supervised training of the model by manually labeling some images and combining a large amount of unlabelled data.
The difficulty in interpreting end-to-end models yields certain limitations when we put this type of computer-aided diagnosis algorithm into practice. Because it is impossible to know the basis for the model to make judgments, the only reliable way to manually perform a second review is to diagnose again. However, reducing the workforce burden of doctors has not been achieved, and computer-aided diagnosis has not achieved its intended effect. Therefore, to improve this situation, we integrate the model analysis algorithm after completing the training model. This type of algorithm can label the key information on the image that the model relies on to make a particular judgment. This method allows doctors to perform a second review when the model is applied, which improves the fidelity of the diagnosis results given by the machine. Also, this method is convenient for researchers to analyze the reasons for the errors of the model to decide the direction of subsequent improvement. The model analysis algorithm is based on model gradient analysis [24] that considers that the more critical information is, the more likely its small changes will affect the results, making the gradient smaller. Conversely, the more unimportant the information, the less likely its change will affect the result, making the gradient smaller. We apply this algorithm to the proposed disc herniation diagnosis algorithm. When the model yields a positive judgment result, the vital information is marked concurrently in the form of a heat map.
In summary, the existing algorithms for automatic diagnosis of lumbar disc herniation mainly have the following two limitations:
-
1.
Most methods focus on processing central sagittal lumbar MRI images and use target detection to achieve an automatic diagnosis, while few use mid-layer transverse lumbar MRI images. MRI is a three-dimensional scanning data that can take multilayer images from the sagittal plane and intercept cross-sectional multilayer images. Most previous studies on this issue only used MRI single sagittal plane images without effectively utilizing the information in MRI three-dimensional data.
-
2.
The interpretability of the model. The model only provides the classification results without providing any judgment basis, which makes it difficult for doctors to make a second confirmation of the results generated by the computer.
Due to the high cost of labeling data, labeling training data is a general difficulty in applying computer technology to the medical image. They are combined with the limitations of existing automatic diagnosis algorithms for disc herniation.In this paper, on the one hand, a two-stage semi-supervised classification model is used to extract comprehensive semantic information to judge the intervertebral disc morphology by taking multilayer intervertebral disc cross-sectional scan images as input, which reduces the quantity requirement and complexity of the labeled data. On the other hand, The interpretability analysis algorithm of Grad-CAM (Gradient-weighted Class Activation Mapping) can make the judgment of the classification model visualized and displayed as a thermal map. To apply the Grad-CAM algorithm to the model proposed in this paper, we make some improvements. The main contributions of this paper are summarized as follows:
-
1.
This paper proposes a novel model structure that allows the model to simultaneously extract features from transverse axial MRI images of three sections of each disc. To alleviate the difficulty of insufficient labeled training data, this paper uses a two-stage paradigm of semi-supervised learning. In the first stage, the method of comparative self-supervised learning is used to train on the unlabeled dataset, In the second stage, general supervised learning is used to train on the manually labeled training set.
-
2.
To compensate for the interpretability problem caused by the black-box nature of the end-to-end network model, this paper uses a model visualization method based on feature map gradient to display abnormal regions in lumbar MRI images in the form of thermal maps, making the results of automatic diagnosis easier to reference.
-
3.
In this paper, we produce a classification dataset of lumbar disc herniation by manual annotation based on publicly available lumbar MRI images and diagnostic reports. This dataset fills the shortage of data resources in the research field of automatic lumbar spine diagnosis algorithms and provides the primary conditions for developing subsequent algorithm research. The dataset is made public to facilitate peer verification and further research.
The remainder of this paper is structured as follows: Sect. Relevant work introduces relevant work, including Two-stage semi-supervised Learning Paradigm, Contrast Self-Supervised Learning, and Interpretability Analysis Algorithms. Section Design of themodel introduces design of the model, including unsupervised pre-training stage, supervised fine-tuning, and generation of heatmaps. Subsequently, experimental procedures and evaluation data are provided in Experiments. Lastly, the key findings of the study are summarized in Conclusions.
Relevant work
Two-stage semi-supervised learning paradigm
The constructed dataset generally consists of two parts: samples and labels. For supervised learning, each sample should correspond to at least one label. The data set used in the complete training process of semi-supervised learning will contain both unlabelled and labeled samples. To complete the classification task, we logically divide the supervised learning process into two steps: feature extraction and feature classification. This feature extraction step can be performed by human design, which was a common method before deep learning became popular. This step can also be performed automatically by deep neural networks. Feature classification aims to infer the label corresponding to the new sample feature through the paired data of the feature and the label in the existing dataset. The conditional probability is being inferred \(P(\text {label}|\text {feature})\). Feature classification can be achieved by multilayer perceptrons or support vector machines. The end-to-end model based on the deep neural network is equivalent to not distinguishing these two steps artificially and composing them as a whole, where all steps are performed “in a black box”.
Contrastive feature extraction and feature classification are two primary procedures. The key to feature extraction is to extract features with semantic value, which is relatively complicated and is one of the reasons why human-designed features have been used until deep learning methods became widespread. Feature classification will be relatively simple if feature extraction is sufficiently good and contains sufficient key semantic information. A simple support vector machine or a perceptron consisting of a shallow neural network will perform well. Therefore, in deep learning, many computer resources are used to extract better features.
The larger the number of labels, the closer the classifier results will be to the actual situation. Although labels are necessary for feature classification, feature extraction does not necessarily require the participation of labels. A typical example is the tf-idf statistical method in the field of natural language processing [25]. To classify the topic of an article, a frequently used method of constructing features is tf-idf (term frequency-inverse document frequency), which consists of tf and idf. Moreover, tf is also known as term frequency, which is the frequency of a specific word appearing in a specific article; idf, also known as inverse document frequency, is the ratio of the number of articles with a specific word in the entire corpus to the total number of articles. This feature extraction method for articles has no label participation; the primary idea is to use the statistical information of an article in the entire corpus as a feature.
In the training process of the classification model, we consider that feature extraction will consume more resources, and feature extraction does not necessarily require labels. Therefore, we use a large amount of unlabelled data for feature extraction learning and then use labeled data for feature classification learning. Hopefully, we can reduce the amount of labeled data required, thereby reducing the cost of building the dataset; this is the primary idea behind two-stage semi-supervised learning.
Therefore, the two-stage semi-supervised model consists of a feature extractor and a feature classifier. The feature extractor is a deep neural network because the feature extraction task is more complex than classification and should use the solid fitting ability of deep neural networks to complete. Feature classifiers can use multilayer perceptrons or support vector machines.
Contrast self-supervised learning
The feature extractor aims to extract key semantic information from input samples. A classic example is the autoencoder [26]. Autoencoders belong to the encoder-decoder paradigm, which includes an encoder and a decoder, as shown in Fig. 1. The encoder is responsible for mapping an original input image into a n-dimensional feature vector, represented by \(f_E:{\mathbb {R}}^{W\times H} \rightarrow {\mathbb {R}}^n\). The decoder is responsible for mapping the above n-dimensional feature vector into an image of the same size as the input, represented by \(f_D:{\mathbb {R}}^n \rightarrow {\mathbb {R}}^{W\times H}\). The feature vector output by the encoder is the vector representation of the input sample, which is the result of feature extraction as well. To ensure that the key semantic information of the input image can be contained in the feature vector, the image output by the decoder should be as identical as possible to the original image. The image output by the decoder is also called the reconstructed image. Therefore, the training goal of the autoencoder is to optimize the parameters \(f_E\) and \(f_D\) so that the difference between the input image and the reconstructed image is as small as possible.
Typical structure of an autoencoder [26]
For feature extraction, autoencoders are a more general method. However, the feature vector extracted by this method strongly considers the information integrity of the input image at the pixel level and lacks practical semantic information. We consider that the training goal of the autoencoder is to reconstruct the image as close as possible to the original input image at the pixel level. Therefore, the resulting feature vectors tend to retain the low-frequency information that represents the overall structure in the image, such as the background, while discarding the high-frequency information that represents the details, such as texture. This method is more like the compression of information than the extraction of semantic information. However, for classification tasks, high-frequency information representing details is often an essential basis for making judgments and is even more critical for medical images. In most cases, when given a specific diagnostic task, different medical image samples are similar in the overall structure. The differences between the samples are primarily reflected in some details, which are often a vital basis for diagnosing. Taking the lumbar intervertebral disc as an example, some localized high signal points in MRI images are often the characteristics of annulus fibrosus tears. Therefore, autoencoders are not the best choice for medical image diagnosis problems.
If we think about the feature extraction task differently to facilitate classification, so the extracted features should be able to reflect the semantic difference between samples as much as possible. For example, the shape of the intervertebral disc, the relative positional relationship with the dural sac, etc. We can ignore the difference in the representation of the same semantics, such as the image’s contrast and the camera’s angle. The feature extraction method designed based on this goal is self-supervision [21, 22]. As shown in Fig. 2.
First, we consider the case with only two input images, A and B. We define a set of random transformation operations \(f_T:{\mathbb {R}}^{W\times H} \rightarrow {\mathbb {R}}^{W\times H}\) that can be applied to the input image. \(f_T\) consists of a series of transformation operations. The definition can vary according to the real situation and only requires that most semantic information will not be lost due to the transformation operation. We can operate with the following transformations: (1) random crop; (2) random rotation; (3) random flip; (4) random change of brightness or contrast; and (5) random Gaussian blur. So limiting the degree of random transformation within a certain range is necessary to train the model more efficiently. If the degree of transformation is too drastic, the original semantic information will be damaged. If the degree of transformation is too small, the model cannot be effectively trained. Input images A and B are thus transformed twice by \(f_T\). Due to the randomness of \(f_T\), two transformed images will be obtained. After two transformations, A becomes \(A_1\) and \(A_2\), and B becomes \(B_1\) and \(B_2\).
We define an encoder \(f_E:{\mathbb {R}}^{W\times H} \rightarrow {\mathbb {R}}^n\) using a deep neural network, which can map two-dimensional images to semantic feature vectors. \(A_1\), \(A_2\), \(B_1\), and \(B_2\) can be mapped to their respective semantic feature vectors by the encoder \(f_E\). This deep neural network is a good choice for using the ResNet-50 network architecture for the encoder. The SimCLRv2 model proposed by Chen et al. [22] uses ResNet-50 and three fully connected layers to form an encoder. The role of the fully connected layer is to map the semantic feature vectors to a smaller dimension.
We look forward to achieving the goal of ignoring the difference in the representation of the same semantics; it is necessary to make \(f_E(A_1)\) and \(f_E(A_2)\) as similar as possible, as well as \(f_E(B_1)\) and \(f_E(B_2)\). The vector inner product is often used in machine learning to express the degree of similarity between two vectors; thus, the above goal can be achieved by simultaneously maximizing \(\left\langle f_E(A_1),f_E(A_2) \right\rangle \) and \(\left\langle f_E(B_1),f_E(B_2) \right\rangle \). However, to prevent the neural network from mapping all inputs to one point, the feature vector is also required to achieve the above goal of reflecting the semantic difference between samples as much as possible. We also need to make the difference between the feature vectors representing different semantics as large as possible. Therefore, the training target of the model also should minimize \(\left\langle f_E(A_1),f_E(B_1) \right\rangle \), \(\left\langle f_E(A_1),f_E(B_2) \right\rangle \), \(\left\langle f_E(A_2),f_E(B_1) \right\rangle \), and \(\left\langle f_E(A_2),f_E(B_2) \right\rangle \). To solve this optimization problem, we can use a description method of the classification problem to design a loss function. A total of 4 feature vectors are involved in this problem: \(f_E(A_1)\), \(f_E(A_2)\), \(f_E(B_1)\), and \(f_E(B_2)\). For \(f_E(A_1)\), there should be one and only one of the other three feature vectors that express similar semantics, which is \(f_E(A_2)\). Therefore, \(f_E(A_1)\) can be classified, which allows one of the other three feature vectors to express similar semantics with \(f_E(A_1)\). The softmax function can be used to describe the result of the classification prediction of a pair of feature vectors. For example, the probability that \(f_E(A_1)\) and other feature vectors express similar semantics can be expressed as:
where \({\mathcal {P}}_{A_1A_2}\) represents \(f_E(A_1)\) and \(f_E(A_2)\), which could express similar semantics probability, while the others are the same. \(\tau \) is a commonly used hyperparameter in the softmax function that is typically referred to as temperature and is used to adjust the absolute value of each exponential part. This hyperparameter prevents the softmax function from being unavailable due to the order of magnitude of the exponent being too large or too small. The true labels should be \({\mathcal {P}}_{A_1A_2}=1\), \({\mathcal {P}}_{A_1B_1}=0\), and \({\mathcal {P}}_{A_1B_2}=0\). The cross-entropy loss can be used as the objective function for model training:
There is no doubt that these formulae describe the case where only two input images are considered. If the feature extractor can obtain the desired effect on all input images, it should be trained with a large amount of unlabelled image data.
The set of all data in the training set is S, and the batch size of batch training is n. Each step in the training process randomly selects n images from S as input, and these n images form the set B used in the training step, \(|B|=n\). If the above formula (1)–formula (4) is extended to the case of n images, there will be 2n transformed images. We thus let \(m_i(1 \leqslant i \leqslant 2n)\) denote the i transformed image, where \(m_{2h-1}\) and \(m_{2h}\) are the h original input image two transformations. We also let \(x_i=f_E(m_i)\) and \(x_i\) represent the semantic feature vector of \(m_i\) after neural network mapping. Therefore, the probability prediction value of the pair of feature vectors \(x_i\) and \(x_j\) expressing similar semantics can be expressed as:
The optimal training objective of the neural network is expressed as:
where \({\mathbb {E}}\) represents expectations and \(\mathbb {1}_{ \left\{ \varPhi \right\} }\) represents an indicator function, and \(\varPhi \) is a proposition. When \(\varPhi \) is true, \(\mathbb {1}_{ \left\{ \varPhi \right\} } = 1\); when \(\varPhi \) is false, \( \mathbb {1}_{ \left\{ \varPhi \right\} } = 0\). \(\lceil \cdot \rceil \) means round up. A feature extractor can be obtained by training a neural network with a gradient descent algorithm.
A complete neural network is not always required when using a trained neural network as a feature extractor. Thus, we can choose the output of one of the layers as the output of the feature extractor because, in the trained neural network, the output of each layer contains semantic features. However, the complexity of the features expressed by different layers is different [27]. The SimCLRv2 model [22] consists of a ResNet-50 network and three fully connected layers. After training, the last two fully connected layers are discarded. The output of the first fully connected layer is used as the output of the feature extractor.
Interpretability analysis algorithms
The external performance of end-to-end convolutional neural networks can be viewed as a black box. When completing a classification or detection task, people generally only care about the input and output, and it is unclear how the logic inside the network operates. Although an unexplainable model will generally not affect its daily use, it will have many hidden dangers in medical imaging research.
First, researchers that train the models cannot determine the models’ reliability. Data leakage can occur during model training, which means that during data collection, some features are highly correlated with the output of the model. However, these characteristics are not the basis for correct judgment. For example, in a study, a neural network is required to predict whether a male patient has prostate cancer. In the dataset used, most samples with prostate cancer are collected from patients who have undergone prostate surgery, while the healthy patients do not undergo surgery. Whether patients have received surgery becomes the basis for the judgment. Thus, models can quickly achieve high accuracy, but this feature does not serve as a basis for diagnosis, which is the mistake of considering consequences as the causes. The introduction of this feature is a mistake caused by ill-consideration in sample collection. If the logic of model judgment is not analyzed, it is possible to ignore this problem and put a meaningless model into use by mistake.
The second point is the review of the results by model users. We consider that the current deep learning technology cannot wholly replace the diagnoses of human doctors. The purpose of any current diagnostic algorithm model is to aid in diagnosis; thus, a review of the results by human doctors is necessary. If the model is treated as a complete black box, the only way to double-check its results is to perform the diagnosis again. Thus, doctors’ efficiency improvement through computer-aided diagnosis will be limited.
The CNN (Convolutional Neural Network) in Fig. 3 represents the neural network to be analyzed. Taking the t classification problem as an example, we input an image that is to be analyzed into the neural network for the next propagation. There will be t neurons in the output layer, and their respective activation values will affect the prediction result. The neuron activation value of the c-th category is \(h^{(c)}\); the activation value of each neuron will usually be processed by the softmax function and become a probability value, indicating the probability that the input image belongs to each category, where \(h^{(c)}\) is the value before the softmax function. We extract the feature map output by the deepest convolutional layer from the to-be-analyzed neural network and let the k channel of the feature map be \(A^{(k)}\). Next, we calculate the gradient of \(y^{(c)}\) with respect to each activation value in the feature map, where the gradient of \(h^{(c)}\) with respect to the k channel of the feature map is \( \dfrac{\partial h^{(c)}}{\partial A^{(k)}}\). In this step, we perform calculations using the backpropagation algorithm, and the obtained gradient tensor has the same dimension as the feature map. We use a global average pooling algorithm and calculate the weight value of each channel based on all gradient values. Global average pooling calculates the global average of each channel in the gradient tensor and obtains the weight of each channel in the feature map affecting the c-th category. The weight value of the k channel’s influence on the c-th category can be expressed as:
where u and v are the height and width of the deepest feature map, respectively. Thus, we can obtain the weight of each channel of the feature map on the c-th category. After the channels of the feature map are weighted and summed with the corresponding weight values, we can obtain a heat map representing each region’s importance in the space for the judgment result by the c-th category. This weighted summation is processed using the ReLu function to limit its value to [0, 1], which represents the importance ratio of each region; the final heatmap of the c-th category is expressed as:
The heatmap obtained by formula 8 has the exact resolution \(u \times v\) as the deepest feature map. This heatmap should be upsampled to the same resolution as the original input image for viewing convenience. Most upsampling algorithms will work well, and it is common to use bicubic interpolation for better perception.
Design of the model
In this study, we use a two-stage semi-supervised classification model to classify the morphology of the intervertebral disc. The shape of the intervertebral disc is divided into two categories: normal and abnormal. Abnormal conditions included intervertebral disc herniation, intervertebral disc bulge, herniation, and annulus fibrosus tear. The overall structure is shown in Fig. 4.
In the unsupervised pre-training phase, we use the unlabelled sample set to train the feature extraction network. Each unlabelled sample consists of a transverse-axial MRI image of the lumbar spine, whose size is \(W \times H\). After the entire feature extraction network training is completed, all fixed feature extraction network parameters remain unchanged and enter the supervised fine-tuning stage. The dataset in the supervised fine-tuning phase uses an annotated dataset. Each of these samples consists of three transverse-axial MRI images of the lumbar spine, which are obtained from the MRI-scanned images of three adjacent cross-sections in the center of the same intervertebral disc. The input sample size is \(W \times H \times 3\). The feature extraction network processes the samples to generate corresponding semantic feature vectors. The semantic feature vector goes through a fully connected layer and is mapped to the prediction result. The technical design details of the two stages are described below.
Unsupervised pre-training stage
The feature extraction network training needs to be completed in the unsupervised pre-training stage. Because the feature extraction network should extract the high-level semantic features of the sample, it is generally constructed using a deep neural network. Considering that deep neural networks are accompanied by gradient disappearance or gradient explosion problems, the network structure represented by RestNet [28] is more commonly used. The SimCLRv2 [22] model also uses RestNet-50 as the basic feature extraction network structure. These methods have only a few subtle differences within their basic structures.
In the unsupervised pre-training stage, the deep neural network consists of a ResNet50 network followed by four fully connected layers. After the transformation function processes the input image, it is input into the neural network. The output of the last fully connected layer is a semantic feature vector, while the neural network is then trained by the contrastive self-supervision method described in 2.2. The overall training process is shown in Fig. 5.
Unsupervised pre-training stage training process. Considering two input images as an example: (a) each input image is processed twice by a random transformation function, and each obtains two transformations; (b) all transformations are paired as a group and then input into the feature extraction network, while each transformation will generate a semantic feature vector; and (c) the two semantic feature vectors of each group are used to predict the semantic similarity; thus, the loss can be obtained, and training will minimize the loss
First, we define a random transformation function \(f_T:{\mathbb {R}}^{W\times H} \rightarrow {\mathbb {R}}^{W\times H}\). \(f_T\) takes a lumbar spine MRI image on the transverse axis as input, and each time, a transformed image is randomly generated according to the specified rules. The batch size used in the unsupervised pre-training phase is n, meaning that each training step randomly selects n images from the dataset S to form a set B. Each image in the set B is transformed by \(f_T\) twice, generating 2n transformed images. \(m_i(1 \leqslant i \leqslant 2n)\) represents the i transformed image, where \(m_{2h-1}\) and \(m_{2h}\) are the two images of the h-th original input image after transformation. All 2n transformations are grouped into pairs and sent into the neural network \(\hat{f_E}:{\mathbb {R}}^{W\times H} \rightarrow {\mathbb {R}}^{d}\). The neural network \(\hat{f_E}\) maps each image to a d-dimensional semantic feature vector. Each set of inputs will generate 2 semantic feature vectors. We let \(x_i\) denote the ith semantic feature vector, where \(x_{2h-1}\) and \(x_{2h}\) are the two semantic feature vectors corresponding to the h-th original input image. Then, we calculate the probability that the two vectors of each group express similar semantics according to the formula 5. Then, we calculate the loss of the set B according to the formula 6 and use the gradient descent method to train the neural network to optimize it.
The neural network \(\hat{f_E}\) used in the unsupervised pre-training stage is not the final feature extractor. The end of the neural network contains four fully connected blocks, whose structure is shown in Fig. 6. The only difference between the last fully connected block and the first to third fully connected blocks is that no ReLu activation function is used. The retention probability of the dropout layer is dr. The feature extractor \(f_E\) used in the subsequent supervised fine-tuning phase discards the last three fully connected blocks. Thus, with the same input, the output of \(f_E\) takes the output of the first fully connected block of \(\hat{f_E}\) as a result. In addition, the output dimensions of the four fully connected blocks in the neural network designed in this study are all d and \(d=256\). The output of each layer is thus a 256-dimensional vector.
To design the random transformation function, we need to increase the magnitude and randomness of the transformation as much as possible to retain most of the key information. We use a series of transformation operations to compose a random transformation function in the following order: (1) random rotation; (2) random cropping; (3) random flip; (4) random change of brightness & contrast; and (5) random Gaussian blur.
Random rotation and random cropping are two coupled operations. The following constraints are required: (1) The cropped area should have overlapping parts in the two random transformations of the same image; (2) The overlapping area should have a high probability of being located in the center of the original image; (3) In the two random transformations of the same image, the rotation center point of the random rotation operation is the same, and each random cropping area should include the random rotation center point; (4) During rotation, a 0-pixel filling area may be generated around the image, while random cropping should avoid these 0 pixels; (5) The aspect ratio of the random cropping area in the original image should be within [3/4, 4/3]; (6) The proportion of the random cropping area in the original image should be larger than \(p_{crop}\); Because the training goal of contrastive self-supervision is to make two transformations of the same image express similar semantics, the two transformations should contain standard information. The information contained in each cannot be much less; thus, constraints (1), (5), and (6) in this study can limit the clipping area. Conversely, the rotation operation may interfere with the implementation of the constraint that the two crops have overlapping regions. If the center points of the two rotations are different, the positions of the regions containing standard information will also be different. Therefore, constraint (3) is used to eliminate this interference. Also, we consider that most essential information in the transverse-axial MRI images of the intervertebral disc is located in the image’s central region. Introducing too many regions near the edges could make the neural network pay attention to less important information; however, we cannot neglect this information thoroughly. Therefore, constraint (2) increases the probability of extracting key information. Finally, the rotation operation may introduce random noise information, which is 0-pixel padding around the image. The rotation of a rectangular image will render some parts of the area outside the rectangle, and the missing part inside the original rectangle will be filled with 0 pixels. To exclude these noises, constraint (4) is introduced accordingly.
The implementation methods of random rotation and clipping satisfying the above constraints are as follows. First, we randomly select a point in the two-dimensional space where the image is located as the “operation center point” \(O(x_c, y_c)\). The distribution of points O follows a two-dimensional Gaussian distribution. We let the random variable \(s \sim {\mathcal {N}}(\mu , \varSigma )\), where:
Operation centre point \(O(x_c,y_c)\) is expressed as:
s is limited to [0, 1], which is equal to 1 when it is greater than the upper limit and equal to 0 when it is less than the lower limit. The coordinates of point O will be rounded to integers.
Both random rotations of the same image take the operation center point \(O(x_c, y_c)\) as the rotation center. The angle of rotation \(\phi \) obeys a uniform distribution within \([-\theta ,\theta ]\), which is \(\phi \sim U(-\theta ,\theta )\). When \(\phi >0\), it rotates anticlockwise; when \(\phi <0\), it rotates clockwise. To alleviate the jagged texture caused by the rotation, we use the bilinear interpolation method to realize the interpolation processing during rotation.
This area can be determined by identifying the coordinates of the upper left corner and the bottom right corner of a cropped region. The upper left point \(P_{ul}\) is a point randomly selected from the upper left of the centre point O with equal probability, which is \(P_{ul} \in \{ (x,y)|0<x<x_c,0<y<y_c \}\). The point \(P_{br}\) is a point randomly selected from the bottom right of the centre point O with equal probability, which is \(P_{br} \in \{ (x,y)|x_c<x<W,y_c<y<H \}\).
To ensure that constraint (4) is met, we let the four endpoints of the clipping area not be located in the 0-pixel filled area; thus, the four endpoints should be checked. For any point (x, y) to be checked, we can imagine that the point around the point O rotates in the opposite direction of the previous random rotation, which is the angle \(-\phi \), which can be calculated as follows:
Then, we check \((x',y')\). If \(0\leqslant x'\leqslant W\) and \(0\leqslant y'\leqslant H\), the to-be-checked point (x, y) is not located in the 0-pixel filled area. Otherwise, checkpoint (x, y) is located in the 0-pixel filled area, and the clipping area does not meet the requirements.
Additionally, we need to check the length and width of the cropped area according to constraints (5) and (6). If the cropped region does not meet any of the constraints (4),(5), or (6), it need to be randomly generated again. There is typically an upper limit on the number of retries. If the upper limit of the retrying number is reached while the transformation result that conforms to the constraints cannot be obtained, the rotation and clipping transformations are skipped. In this study, the maximum number of retries is set to 100. The cropped image is upsampled to the same resolution \(W \times H\) as the original image. The two transformations of the same input image use the same operation center point to select the cropping region.
The random flip operation only includes left and right flips, and the probability of flipping is 50%. This operation is relatively simple and will not be repeated in this study.
The random change in brightness makes all pixel values in the image scale up or down by a random factor. The random coefficient of the transformation is \(\kappa _{bright} \sim U(0.6,1.4)\). Then, the values of all pixels in the image are multiplied by \(\kappa _{bright}\). Thus, the input image is processed using the following function:
The random change of contrast makes the image’s histogram expand or shrink horizontally according to a random coefficient. The random coefficients of the transformation are \(\kappa _{contrast} \sim U(0.6,1.4)\), and then the input image is processed using the following function:
where \({\bar{I}}\) is the average value of all pixel values in the image because the image in this study is a grayscale image with only one channel. \({\bar{I}}\) can be calculated as:
The Gaussian blur algorithm uses a Gaussian convolution kernel to convolve the input image. We fix the convolution kernel size in the horizontal and vertical directions as 0.05W and 0.05H. The convolution kernel \(\sigma \) value of the random Gaussian blur is a random variable. \(\sigma \) follows a uniform distribution: \(\sigma \sim U(0.1,2.0)\).
When the feature extraction neural network is trained via the above process, a validation set consisting of a small number of samples randomly selected from the unlabelled sample set in advance will be used concurrently. The accuracy of the predicted results is used as the test metric. After a training period, the accuracy no longer continues to improve, and the unsupervised pre-training phase can be ended. We use this resulting feature extractor for the next step of supervised fine-tuning.
Supervised fine-tuning
As shown in Fig. 4, the samples in the supervised fine-tuning stage consist of 3 lumbar spines transverse axial MRI images corresponding to 3 cross-sections in the center of the intervertebral disc. Each sample has a label that indicates whether the disc has an abnormal or normal morphology. The supervised fine-tuning phase aims to train a classifier to distinguish intervertebral discs. Therefore, it is necessary to input the information of all the cross-sections together into the classifier. The images corresponding to the three sections will pass through the feature extractor to obtain three semantic feature vectors. We connect the three semantic feature vectors to obtain the semantic feature vector of the intervertebral disc sample. The flowchart for this training phase is shown in Fig. 7.
During supervised fine-tuning, the classifier we use to classify the feature vectors is a neural network consisting of only one simple, fully connected layer. The input of the fully connected layer is a feature vector of \(3 \times 256\) dimensions, while the input is one-dimensional (scalar). No other layers, such as random deactivation or batch regularization layers, are required before or after the fully connected layer. The output value of the fully connected layer will be mapped to a value between [0, 1] through the sigmoid function, which is required to represent the probability that the sample is positive. Throughout the process, all parameters in the feature extractor are fixed and maintained in the state when the pre-training is completed. Only the parameters in the fully connected layers will change with training.
We consider that there may be some meaningless noise information around the edges of the input image. We apply a simple data augmentation strategy to the input image during training to give the model better generalizability. This data augmentation strategy performs random transformations on the input image. The transformation strategy we use in this stage of training is different from that used in the unsupervised pre-training stage and does not require overly complex transformations. Its primary purpose is only to remove a portion of the surrounding area of the input image but it cannot lose the content of the primary part of the image. This study uses a specific cropping strategy to transform the image.
We randomly select a point \(P_{ul}\) as the upper-left corner of the cropped area and a point \(P_{br}\) as the bottom-right corner of the cropped area. For the upper left point, \(P_{ul} \in \{(x,y)|0<x<0.25W,0<y<0.25\,H \}\), we randomly select a point in this area with equal probability as \(P_{ul}\). For the lower right corner point, \(P_{br} \in \{(x,y)|0.75W<x<W,0.75H<y<H \}\), we randomly select a point in this area with equal probability as \(P_{br}\). These two points can determine a rectangular area. Therefore, we crop the three sections in the same sample according to this area and then upsample to the resolution \(W \times H\) of the input image.
Generation of heatmaps
For positive samples, we can generate a heat map showing the location of the key information for the model to make a positive judgment. This location can be used to assist doctors in judgment and review while negative samples are not generated. This method is based on Grad-CAM with improvements and adaptations. The general Grad-CAM algorithm is introduced in the Interpretability analysis algorithms section, and the algorithm obtains a heatmap for each input image.
In the forward propagation process of the inference process, the feature map output by the deepest convolutional layer in the RestNet network is denoted as A, which contains all the feature maps of the three images as input. Assuming that each input image corresponds to n feature maps, the k feature map is \(A^{(k)}\). Then, we use the backpropagation algorithm to calculate the gradient of the activation value in this layer with respect to the classifier output value h before the sigmoid function, denoted by G, where \(G=\dfrac{\partial h}{ \partial A}\). According to the classic Grad-CAM approach, we should use the global average pooling algorithm to calculate the weight value of the gradient G and then weight the feature map according to the weight value. Finally, we use the ReLu function to obtain the heatmap, such as formula (7) and formula (8). However, first, we consider that the element values in the gradient tensor G have both positive and negative values. For the gradient values with the same absolute values, both positive and negative should have the same importance. Therefore, to prevent positive and negative values from canceling each other, we use |G| to calculate the weight value of the feature map. Next, we generate a corresponding heatmap for each input image because the classifier built into this study takes MRI scan images of 3 intervertebral disc sections as input. Additionally, we present the weight ratio of each of the three images; thus, the weight of each feature map is calculated as:
where \(\sigma (\cdot )\) represents the standard deviation. The formula (17) is used to normalize the calculated weight value. The weight of each feature map is obtained via the softmax function. Then, we weighted the n feature maps corresponding to each image by their weight values. We finally normalize each heatmap separately, limiting it to [0,1] to obtain a standard heatmap. The calculation occurs as follows:
where \(H^{[a,b]}\) represents the tensor composed of channels a to b of H. The max function aims to find the maximum value in the entire tensor.
Experiments
Construction of the dataset
The raw data we used originated from Sudirman et al.’s published lumbar spine MRI data [29], which included sagittal and transverse axial lumbar spine MRI scans of 515 patients. Most scanned images are recorded with the patient in the head-first supine position, and the scans are organized in DICOM (Digital Imaging and Communications in Medicine) format. Both sagittal and transverse axial scans have T1- and T2-weighted images. The data for each patient covered scans of the lowest 3–6 intervertebral discs. For the scan results of the transverse axis, each intervertebral disc has 4–5 cross-section scan data. All the cross-sections in the same intervertebral disc are parallel to each other, and the scanning range of the cross-section is a regular quadrilateral area with a side length of 220 mm. The thickness of the cross-sections varies from 3.0 mm to 5.0 mm, and the distance between two adjacent cross-sections of the same intervertebral disc varies from 3.3 mm to 6.5 mm. Each patient is described by a diagnostic report from the radiologist, which uses natural language to describe the patient’s abnormal disc position, abnormal appearance in the image, and diagnostic conclusion. The entire dataset contains 48,345 images, of which there are 17,219 transverse axial images. Most images have a resolution of \(320 \times 320\), and a small part is \(320 \times 310\). All images are grayscale, and each pixel is represented with 12-bit precision.
We can determine that the format of this dataset is non-standardized. Therefore, we organized it into a standardized dataset and manually annotated some data combined with the doctor’s diagnosis report.
The first is the construction of the dataset used for unsupervised pre-training (“the pre-training dataset”), which uses all transverse-axial cross-sectional MRI scans of the intervertebral disc, including T1- and T2-weighted images. We extract all images and include them in the pre-training dataset, and each image is considered a sample. All images are downsampled to a resolution of \(300 \times 300\). After all images are randomly shuffled, we sample 512 images as the validation set. Because the pre-training dataset is task-agnostic, there is no need for testing. Therefore, we do not need a test set; the remaining 16,707 images are used as training sets.
The second is the construction of the dataset used for supervised fine-tuning. The purpose of this dataset is to classify intervertebral discs (“the disc classification dataset”). The construction of this dataset is relatively complex because in reality, radiologists generally only use T2-weighted MRI images of the transverse axis when diagnosing lumbar spine diseases. Thus, the disc classification dataset is constructed using only T2-weighted images. Conversely, in the intervertebral disc classification dataset, one sample corresponds to one intervertebral disc and contains three cross-sectional images of the intervertebral disc. Ideally, these three sections should be in the vertical center of the intervertebral disc. However, considering that the form of the original data set is nonstandard, for example, an intervertebral disc may contain four or five cross-sections of varying numbers, the distance between the cross-sections may not be constant. We use the following strategy when constructing the disc classification dataset to obtain a model that generalizes as well as possible. First, for an intervertebral disc with five cross-sections, take the central three layers, where the second, third, and fourth layers are intervertebral disc samples; for an intervertebral disc with four cross-sections, take the first three layers as intervertebral disc samples, and then take the back three layers as another disc sample. For such a disc, two-disc samples are generated. Second, we only selected the lumbar intervertebral discs as samples and discarded all the data of thoracic and sacral intervertebral discs. In addition, there may be some duplicate data in the original data; thus, deduplication need to be performed when constructing an intervertebral disc classification dataset.
To label the intervertebral disc classification dataset, we use the diagnostic report given by the radiologist as the basis and label each intervertebral disc after manually reading the report. An intervertebral disc will be marked as a positive sample if bulging, herniation, nerve root compression, dural sac compression, or annulus fibrosus tearing are noted. If it is a typical patient or a patient unrelated to the intervertebral disc, such as a patient with lower extremity arthritis, fracture, and other conditions, the intervertebral disc data will be marked as a negative sample.
The annotated disc classification dataset contains 3676 samples, including 1706 positive and 1970 negative samples. After randomly shuffling the intervertebral disc classification data set, 256 positive and negative samples were randomly selected to form the test set, and 64 positive and negative samples were randomly selected to form the validation set.
Experimental environment and training parameters
The programming environment used in this experiment is Python\(-\)3.7.6. We use the CUDA11.3+TensorFlow\(-\)2.3 model to build a neural network. Training is performed using a GTX3090 graphics card. The optimizer uses the adaptive moment estimation optimizer (Adam), the learning rate is lr, and the batch size of batch training is b. Both stages of training will perform L2 regularization on all the weight coefficients in the network, and the regularized weight coefficient is w. The experimental hyperparameters used in the unsupervised pre-training stage are shown in Table 1, and the experimental setting values of the hyperparameters in the supervised fine-tuning stage are shown in Table 2:
The input to the model is a grayscale image of \(300 \times 300\), i.e., \(W=H=300\). The dropout layer is only effective in the unsupervised pre-training stage. However, it has been experimentally verified that the results will be better when the retention probability of the dropout layer is 0. This fact will be explained in detail in the experimental section. \(\theta \) is the range of the random rotation in the random transformation process, and \(p_{crop}\) is the proportion of the randomly cropped area. Both are valid only in the unsupervised pre-training stage. The training continues until the accuracy on the validation set no longer improves and the loss function value of the model no longer decreases.
Model evaluation methods
We primarily use the following metrics to evaluate the performance of the model. Accuracy (ACC) is the proportion of samples whose predicted value is the same as the real value in all samples:
Sensitivity, also known as the true positive rate (TPR), is the ratio of true positive samples to real positive samples:
Specificity, also known as the true negative rate (TNR), is the ratio of true negative samples to real negative samples:
Because the confusion matrix of the binary classifier is dependent on the classification threshold, to exclude the threshold from the evaluation system and to reflect the comprehensive performance of the classifier under all possible classification thresholds, the ROC curve [30] can be used. For an ideal binary classifier, the area under the ROC curve should be 1. We use an indicator AUC (Area Under the ROC Curve) to represent the area under the ROC curve, which can well reflect the overall performance of the two classifiers.
Effect of regularization level on feature extractor performance
In the unsupervised pre-training stage, we use a strategy of L2 regularization and dropout regularization for the model. The weight parameter w of L2 regularization and the retention probability of dropout dr can describe the strength of the regularization level. For a classifier, a suitable regularization level will give the model better generalizability, while an excessive regularization level will lead to underfitting. For the proposed feature extractor, an appropriate level of regularization will allow the feature extractor to extract higher-quality feature vectors.
We set 4 different sets of regularization parameters, keeping the other hyperparameters unchanged, and train four different feature extractors. We train each feature extractor via supervised tuning using the same settings for 200 epochs. The obtained classifiers are tested on the test set, and the respective performances are compared and analyzed. The results are shown in Table 3. The respective ROC curves of the 4 different cases are shown in Fig. 8.
Because the test set in this study is balanced, both accuracy and AUC can comprehensively measure the classifier’s performance. Based on the experimental results, with the improvement of the regularization level, both the accuracy and AUC experienced a process of increasing first and then decreasing. When \(w=0 \,and \,dr=0\) (i.e., no regularization is performed), the accuracy and AUC are not optimal. In the two cases of \(w=1\times 10^{-4},dr=0\) and \(w=1\times 10^{-4},dr=0.25\), the results of the classifier on the test set are nearly identical. There is some chance that accuracy, sensitivity, and specificity are identical. However, the classification performance of the two is similar, while the AUC of the latter has decreased markedly. When a strong regularization level is used, such as \(w=1\times 10^{-4} \,and\, dr=0.5\) in the experiment, the classifier’s performance will decrease markedly. Therefore, choosing appropriate regularization parameters is critical to training feature extractors. In reality, there is no universally applicable optimal setting for the regularization parameter due to certain differences between different datasets. We should fine-tune the model for different datasets because small L2 regular weights and dropout parameters can typically achieve good results. In the model developed in this study, we choose \(w=1\times 10^{-4},dr=0\) as the regularization parameter setting.
Effect of the number of labels
How much the two-stage semi-supervised learning paradigm can reduce the dependence on the number of labels in the training data set and how much the advantage is compared to classic supervised learning are questions worth investigating.
First, we train multiple different classifiers by varying the number of labeled samples used in the supervised fine-tuning phase. All classifiers use the exact same feature extractor. The feature extractor obtained from the unsupervised pre-training stage remains unchanged. In the supervised fine-tuning phase, we randomly sample parts of the data from the disc classification dataset. The multiple classifiers trained in this experiment are randomly selected from the original dataset with 1024, 1536, 2016, and 2528 samples as training subsets. The other training conditions and parameters are the same as those given in Experimental environment and training parameters, and the training lasts for 200 epochs. The original full disc classification dataset contains 3036 training samples, and the four classifiers using the training subset will be compared with the models obtained using the entire training set.
In addition, to facilitate the comparison of the proposed model with the classic supervised learning model, we use the ResNet50 model [28] to train a classifier by fully supervised training on the disc classification dataset with a learning rate of \(1\times 10^{-4}\) for 370 epochs. Other models using the two-stage semi-supervised learning paradigm will be compared.
After testing the above models on the test set, we obtained the accuracy (ACC) and AUC of each model. The data are shown in Fig. 9. The horizontal axis in the figure is the number of labels, which represents the number of labeled data used, while the vertical axis represents the accuracy and AUC index. For the convenience of comparison, we also put the supervised model using ResNet50 into the figure, which is represented by “supv” on the horizontal axis.
As shown in the figure, as the number of labels increases, both the accuracy and AUC of the classifier increase. Concurrently, when the variation in the number of labels is marginal, the performance of the classifier under a certain indicator may also change marginally, such as the accuracy of the two classifiers when the number of labels is 2528 and 3036, and the AUC of the two classifiers when the number of labels is 1024 and 1536. However, when the number of labels increases markedly, a prominent improvement in model performance occurs. This result also shows that the performance of the two-stage semi-supervised learning model has certain stability when using the training subset.
Conversely, the training data used by the trained, supervised model are also the original training set. The trained, supervised model contains 3036 labels, while the semi-supervised learning model in this paper only requires 1024 labels to achieve similar performance. When a semi-supervised learning model is trained on the original training set, the overall performance is much better than that of classical supervised learning. This result indicates that the two-stage semi-supervised learning paradigm can extract effective semantic information from a large amount of unlabelled intervertebral disc MRI data, thereby enhancing supervised classification learning and enabling the classifier to achieve better results.
Classifier heatmap visualization experiment
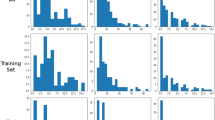
We visualize the heat map of all the data in the test set that are judged as positive samples by the classifier, some of which are shown in Fig. 10. The heat map can indicate the cross-sectional location with more severe degenerative changes for the positive intervertebral disc samples and display it with a higher heat value. The classification basis of the classifier is generally the intervertebral disc area in the image, which indicates that the classifier can read the effective semantic information in the image. For a herniated or bulging intervertebral disc, the heat map can clearly show the location of the herniated or bulged disc, which is worthy of reference.
Conclusions
The medical basis of the automatic diagnosis of lumbar intervertebral disc herniation is to determine the shape of the intervertebral disc. Its standard method in artificial intelligence is to use a supervised method to train an object detection model, which primarily has two difficulties in practice. The first point is the high cost of labeling data. The object detection model could be clearly marked with a large number of bounding boxes and the corresponding intervertebral disc shape categories. Generally, at least thousands of labeled samples are required to train a more usable model. The second point is the difficulty in interpreting the end-to-end model. Doctors will spend more time reviewing the model results when an uninterpretable model is used as a computer-aided diagnosis model. It is also not easy for model researchers to determine the rationality of such a model’s results. To address these two difficulties, we use a two-stage semi-supervised classification model, consider the multilayered intervertebral disc cross-sectional scanned images as input, and extract comprehensive semantic information to determine the intervertebral disc shape, which reduces the quantity and complexity of labeling data. Conversely, we improve and adapt the Grad-CAM interpretability analysis algorithm so that it can be applied to the proposed model to visualize the judgment basis of the classification model in the form of a heatmap.
This study annotates public MRI data to create training and testing datasets. The trained model can achieve an accuracy of 87.11% on the test set, a sensitivity of 87.50%, a specificity of 86.72%, and an AUC index of 0.9487.
After describing the effect of the regularization level on the performance of the feature extractor, we notice that a suitable regularization level enables the feature extractor to extract higher-quality semantic feature vectors and the classifier to obtain better results. Generally, relatively small regularization parameters can achieve better results. By training the model with subsets with different numbers of labels, the two-stage semi-supervised training model can achieve better performance for the disc MRI image classification task. Even with few labels, the resulting classifier performance remains stable. It is possible to achieve comparable levels with classical supervised models using only approximately one-third of the number of labeled training subsets. After visualizing the data in the test set in the form of a heat map, the classification basis of the classifier can be clearly shown. The position of the intervertebral disc with more severe degenerative changes is also shown with a higher heat value.
The diagnosis model of intervertebral disc herniation in this study can thus produce favorable results that can play a critical role in auxiliary diagnosis to a certain extent. The proposed model can reduce the burden on radiologists, improve the efficiency of diagnosis, and provide a reference for follow-up research. Simultaneously, there are still some limitations in the study of computer-aided diagnosis of lumbar disc herniation in this paper, which is worthy of further study:
-
1.
This paper’s auxiliary diagnostic model of lumbar disc herniation cannot specifically distinguish the specific categories of lesions. Future work will refine the categories of degenerative changes so that the model can distinguish the specific categories of various degenerative changes such as bulging, protrusion, prolapse, Schmoy’s nodule, and annulus fibrosus tear.
-
2.
Try to use other semi-supervised algorithms to design the model, compare its performance, and optimize the model proposed in this paper. A sufficient number of MRI images of typical cases were collected from multiple hospitals to train the model to improve its accuracy and practicability.
-
3.
In this paper, sagittal and transverse images were selected artificially when establishing the data set. In the future, MRI scan data in complete DICOM format will be used as the input of the diagnostic system, and all data will be analyzed without artificial separation.
-
4.
In the future, we need to study the relationship between the morphological manifestations on imaging and the appropriate treatment plan. It is expected that through the automatic diagnosis algorithm, the proposed treatment plan for patients can be directly given, and the evaluation and implementation of the prevention and rehabilitation plan of lumbar disc herniation can be guided.
References
Waxenbaum JA, Reddy V, Futterman B (2017) Anatomy, back, intervertebral discs
National Institute of Neurological Disorders and Stroke (NINDS) (2008) Low back pain fact sheet, NIND brochure
Fardon DF, Williams AL, Dohring EJ, Reed Murtagh F, Gabriel Rothman SL, Sze GK (2014) Lumbar disc nomenclature: version 2.0: recommendations of the combined task forces of the north American spine society, the American society of spine radiology and the American society of neuroradiology. Spine J 14(11):2525–2545
Haughton V (2004) Medical imaging of intervertebral disc degeneration: current status of imaging. Spine 29(23):2751–2756
Cheng F, You J, Rampersaud YR (2010) Relationship between spinal magnetic resonance imaging findings and candidacy for spinal surgery. Can Fam Phys 56(9):e323–e330
Ebrahimzadeh E, Fayaz F, Ahmadi F, Nikravan M (2018) A machine learning-based method in order to diagnose lumbar disc herniation disease by mr image processing. MedLife Open Access 1(1):1–10
Oktay AB, Albayrak NB, Akgul YS (2014) Computer aided diagnosis of degenerative intervertebral disc diseases from lumbar mr images. Comput Med Imaging Graph 38(7):613–619
Zheng H-D, Sun Y-L, Kong D-W (2022) Deep learning-based high-accuracy quantitation for lumbar intervertebral disc degeneration from mri. Nat Commun 13:841
Jamaludin A, Lootus M, Kadir T, Zisserman A, Urban J, Battié MC, Fairbank J, McCall I (2017) Automation of reading of radiological features from magnetic resonance images (mris) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J 26(5):1374–1383
Wang Z, Qin J, Huang J, Wang Y, Li J (2020) Automatic diagnosis of disc herniation based on densenet fusion model. In 2020 8th International Conference on Digital Home (ICDH), pages 294–298. IEEE
Gong H, Liu J, Chen B, Li S (2022) Resattengan: Simultaneous segmentation of multiple spinal structures on axial lumbar mri image using residual attention and adversarial learning. Artif Intell Med 124:102243
Han M, Liu L, Mengzi H, Liu G, Li P (2022) Medical expert and machine learning analysis of lumbar disc herniation based on magnetic resonance imaging. Comput Methods Programs Biomed 213:106498
Kuang X, Pui Yin Cheung J, Wong K-YK, Lam WY, Lam CH, Choy RW, Cheng CP, Wu H, Yang C, Wang K et al (2022) Spine-gflow: A hybrid learning framework for robust multi-tissue segmentation in lumbar mri without manual annotation. Comput Med Imaging Graph 99:102091
Mbarki W, Bouchouicha M, Frizzi S, Tshibasu F, Farhat LB, Sayadi M (2020) Lumbar spine discs classification based on deep convolutional neural networks using axial view mri. Interdiscip Neurosurg 22:100837
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. nature 521(7553):436–444
Cheng J-Z, Ni D, Chou Y-H, Qin J, Tiu C-M, Chang Y-C, Huang C-S, Shen D, Chen C-M (2016) Computer-aided diagnosis with deep learning architecture: applications to breast lesions in us images and pulmonary nodules in ct scans. Sci Rep 6(1):1–13
Gargeya R, Leng T (2017) Automated identification of diabetic retinopathy using deep learning. Ophthalmology 124(7):962–969
Wang D, Khosla A, Gargeya R, Irshad H, Beck AH (2016) Deep learning for identifying metastatic breast cancer. arXiv preprint arXiv:1606.05718
Sarraf S, Tofighi G (2016) Alzheimer‘s Disease Neuroimaging Initiative, et al. Deepad: Alzheimer‘s disease classification via deep convolutional neural networks using mri and fmri. BioRxiv, page 070441
Zhu X, Goldberg AB (2009) Introduction to semi-supervised learning. Synthesis Lect Artif Intell Mach Learn 3:1
Oord van den A, Li Y, Vinyals O (2018) Representation learning with contrastive predictive coding. arXiv preprint arXiv:1807.03748
Chen T, Kornblith S, Swersky K, Norouzi M, Hinton GE (2020) Big self-supervised models are strong semi-supervised learners. Adv Neural Inf Process Syst 33:22243–22255
Chen M, Radford A, Child R, Wu J, Jun H, Dhariwal P, Luan D, Sutskever I (2020) Generative pretraining from pixels. In Proceedings of the 37th International Conference on Machine Learning
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision, pages 618–626
Salton G, Buckley C (1988) Term-weighting approaches in automatic text retrieval. Inf Process Manag 24(5):513–523
Bank D, Koenigstein N, Giryes R (2020) Autoencoders. arXiv preprint arXiv:2003.05991
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. In European conference on computer vision, pages 818–833. Springer
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 770–778
Sudirman S, Al Kafri A , Natalia F, Meidia H, Afriliana N, Al-Rashdan W, Bashtawi M, Al-Jumaily M (2019) Lumbar spine mri dataset, Mendeley Data
Fawcett T (2006) An introduction to roc analysis. Pattern Recogn Lett 27(8):861–874
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This study was supported in part by the key research and development program of Hebei Province, innovative product R & D of wearable lumbar rehabilitation flexible exoskeleton robot under Grant 20371801D, in part by National key research and development program of China under Grant 2019YFB1312500.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hou, C., Li, X., Wang, H. et al. An MRI image automatic diagnosis model for lumbar disc herniation using semi-supervised learning. Complex Intell. Syst. 9, 5567–5584 (2023). https://doi.org/10.1007/s40747-023-00981-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-023-00981-0