Abstract
Purpose of Review
Azole resistance in Aspergillus fumigatus is an emerging public health issue with global distribution and has been linked to use in agricultural and horticultural settings. In 2022, the World Health Organization (WHO) created a fungal pathogen priority list, and A. fumigatus was listed as a critical pathogen. Currently, Africa lacks effective surveillance systems for this emerging threat, mostly due to lack of capacity and diagnostics to determine azole resistance in routine clinical settings. This review aims to address and improve on the current diagnostic tools and future perspective strategies in tackling clinical and environmental antifungal-resistant (AFR) A. fumigatus in Africa. We emphasized on the importance of early diagnosis and misdiagnosis associated with aspergillosis caused by Aspergillus sp., cross talk between clinical and environmental, mode of action and resistance mechanism, collaborative one health approach, and future perspectives for AFR A. fumigatus management strategies.
Recent Findings
Early diagnosis and effective management of invasive aspergillosis are critical. On the continent, very few laboratories routinely conduct antifungal susceptibility testing on Aspergillus species. Where this occurs, it is culture-based in vitro antifungal susceptibility testing. Drug repurposing and the need for a non-culture-based molecular method (PCR) are critical.
Summary
Enhancing promising future perspectives of non-cultured approaches such as whole-genome sequencing, CRISPR/Cas9, and RNAi-mediated technologies to complement the culture-based approach as important strategies to mitigate and overcome emerging issues of AFR A. fumigatus in Africa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The prevalence of fungal infections is increasing and poses a global threat to humans [1]. Over 1.6 million deaths annually are caused by fungal diseases globally, and more than 1 billion people are at extremely high risk from severe fungal infections [2, 3]. There are several reasons for this increase, and the major risk factor for invasive fungal infections is the prolonged survival in immunocompromised patients with diseases such as HIV/AIDS, chronic obstructive pulmonary disease (COPD), tuberculosis (TB), and severe acute respiratory syndrome coronavirus (SARS-CoV-2) [4,5,6]. Fungal infections occur across a range of medical conditions, usually as co-infections or opportunistic infections [7, 8]. Therefore, invasive fungal infections are complicated, which hinders diagnosis and management of immunocompromised patients [9].
The World Health Organization (WHO) developed the first global fungal priority pathogens list (FPPL), which aligns with public health requirements [10]. Recently, the WHO updated the list of fungal pathogens into three priority groups (critical, high, and medium) using a multi-criteria decision analysis (MCDA) approach for use in public health systems in response to fungal infections and antifungal resistance [11•]. The critical group of priority pathogens are Aspergillus fumigatus, Candida albicans, Candida auris, and Cryptococcus neoformans. Therefore, the focus is on invasive acute and subacute systemic causative fungal pathogens with drug resistance or management challenges.
Africa accounts for one fifth of the world’s population that are at risk for contracting fungal infections with limited access to healthcare, harmful environmental factors, and generally poor livelihoods [12]. The high incidence of fungal infections in Africa is a major concern, and not all healthcare systems are able to address this challenge. Data compiled by the Global Fund Action for Fungal Infections (GAFFI) suggests that 47.6 million Africans suffer from fungal diseases, of which 1.7 million suffer from a severe fungal infection [13]. However, these assessments are based on data from only a handful of African countries and likely underestimate actual prevalence. Fungal infections such as bloodstream infections, wound infections, and urinary tract infections are caused by antifungal-resistant (AFR) A. fumigatus pathogen, which have been reported in Nigeria and West Africa [14].
To date, the treatment of invasive fungal infections caused by Aspergillus spp. relies primarily on these antifungals: amphotericin B (polyenes), fluconazole, voriconazole, itraconazole (azoles), and caspofungin (echinocandins) [15,16,17]. However, resistant fungi react quickly to chemical attacks [18], and failed treatment is a common consequence of resistance. This is attributed to an interplay between underlying host immunodeficiencies, antifungal drug properties (drug-target interactions, pharmacokinetics, and pharmacodynamics) [19], biofilm formation [20], and fungal properties (varied cell morphologies, antifungal tolerance, antifungal resistance, genetic mutations, and induced protective mechanisms) [21, 22]. Rapid plasmid-mediated spread of resistance has not been demonstrated in fungi relative to bacteria [23], and such mechanisms need to be investigated.
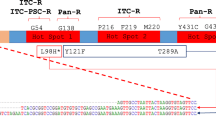
Unfortunately, studies report global resistance to azole, echinocandins, and polyene antifungal drugs in both clinical and environmental fungal strains, which were previously effective against A. fumigatus [15, 24] and C. auris [25, 26••]. Azole drug resistance has severe clinical consequences, with retrospective studies showing a 25% increase in mortality at day 90 in patients with drug-resistant aspergillosis relative to patients with wild-type (WT) infections [27]. In general, the prevalence of azole resistance in A. fumigatus is characterized by the expression-upregulating tandem repeat (TR) mechanisms in the promoter region of the Cyp51A gene through point mutations, which decrease the affinity of azoles for the target protein [28, 29]. Hence, the most common alleles, namely TR34/ L98H and TR46/Y121F/T289A, are associated with high levels of itraconazole and voriconazole resistance [30], both inside and outside clinics [27].
In Africa, emerging high drug-resistant pathogens are gaining clinical importance [26••]. Modern genomic epidemiological methods have been extensively explored and revealed possible eco-evolutionary associations between the environment and increasing clinical resistance of the azole-resistant genotypes A. fumigatus [24] in patients with no history of antifungal treatments. However, ecological “hotspots” have been postulated [18], where both biotic and abiotic conditions allow fungal growth when exposed to azole concentrations below the minimum inhibitory concentration (MIC), creating suitable conditions for adaptation to drug pressure. Therefore, these pathogens need to be screened and ascertained for the burden of outbreak infections if they are to be prioritized for health interventions. In this review, we focused on these priority areas with the aim of outlining current advances and future perspectives regarding key research strategies needed to mitigate the invasive antifungal-resistant pathogen A. fumigatus in Africa.
Diagnosis and misdiagnosis associated with aspergillosis
Aspergillosis is undoubtedly the most common human disease, ranging from superficial to deep-seated and potentially fatal infections [31, 32], for example, otomycosis, onychomycosis, keratitis, chronic pulmonary aspergillosis (CPA), allergic bronchopulmonary aspergillosis (ABPA), saprophytic pulmonary, or sinusoidal aspergillomas, which develop rapidly and are often fatal, especially if diagnosis is delayed or missed [33, 34]. Some of these fungal infections, particularly skin diseases that affect individuals without any pre-existing conditions who live and work in close proximity to certain environmental niches [35, 36].
The morphological diagnosis of aspergillosis is usually obtained by the observation of thin, septate, acute-angled, or dichotomously branched hyphae [37]. Aspergillus is a large genus of about 250 species that has a number of important pathogens. Some pathogenic species are present in culture contaminants, with varying pathogenicity and susceptibility to antifungal agents. The number of these pathogenic species is increasing rapidly due to adaptation to temperature, and their numbers may be currently underestimated [36]. Nonetheless, the rapid and accurate identification of clinical isolates is very important in selecting appropriate antifungal agents.
Diagnostic challenges also occur at the morphological level, as there are some hyaline septate molds that appear to be indistinguishable. For example, Fusarium spp., Scedosporium sp., and Pseudallescheria sp. were diagnosed in patients between histology and culture ranges from 17 to 22% [38,39,40]. The therapeutic importance of this is significant, as there are significant differences between the treatment of aspergillosis and that of mucormycosis (mucormycetes). Molecular methods are more time-consuming than direct microscopy and response to treatment depends on early diagnosis and the use of appropriate antifungal agents and, therefore, considered a diagnostic challenge [41].
Clinical diagnosis of aspergillosis is often challenging due to nonspecific clinical features, and the significance of this is that they are being treated with wrong medicines [42, 43]. Therefore, most lesions do not respond well to therapy and may become chronic or kill the patient. The prevalence of onychomycosis due to Aspergillus sp. varies in different parts of the world. The incidence of onychomycosis by Aspergillus has shown an increase in recent years, representing 34–60% of onychomycosis due to non-dermatophyte molds [44, 45]. Several Aspergillus spp. have been isolated from human nails such as A. fumigatus, A. flavus, A. versicolor, A. niger, A. terreus, A. sclerotiorum, and A. nidulans [44]. Aspergillus onychomycosis infections are often misdiagnosed and targeted with fluconazole leading to treatment failure and chronicity of the disease [46].
Over 3 million cases of chronic pulmonary aspergillosis have been estimated to occur globally each year, and 10 million cases of fungal asthma occur yearly, yet a significant proportion remain undiagnosed in resource-poor communities [3, 47•]. In addition, fungal asthma, caused by airborne fungi such as Aspergillus spp., exacerbates asthma in millions of adults and children [48, 49]. Limited diagnosis or misdiagnosis and poor estimates of disease morbidity result in the true burden of disease not being fully known [50]. In pulmonary aspergillosis, the rate of misdiagnosis due to nonspecific clinical findings and atypical radiological manifestations is up to 73% [51]. Previous study showed that chronic pulmonary aspergillosis (CPA) can be diagnosed as bacteriologically negative pulmonary tuberculosis (TB) [52]. This has been attributed to lack of awareness and limited access to Aspergillus-specific IgG testing and CT imaging in India. In 29 to 45% of aspergillosis cases, ABPA are often confused with pulmonary TB, and patients receive many doses of anti-tuberculosis drugs before diagnosis are performed [53].
Patients with severe COVID-19 pneumonia are susceptible to secondary viral, bacterial, and fungal infections due to diffuse alveolar lung damage and dysregulated immune response [54,55,56]. Early diagnosis of these co-infections is important in order to initiate appropriate antimicrobial therapy [57]. Coronavirus disease (SARS-CoV-2, COVID-19)–associated pulmonary aspergillosis (CAPA) is a syndrome affecting COVID-19 patients with acute respiratory distress syndrome (ARDS) requiring intensive care in intensive care units (ICUs), with incidence rates ranging from 3.8 to 33.3% [58], and thus, these diagnostic variations are attributed to differences in the patient populations and CAPA. The diagnosis of CAPA is complex due to the commonly observed atypical radiological features, the lack of established host factors, and the difficulty in obtaining mycological evidence due to the low sensitivity of serum galactomannan in CAPA [59, 60]. Early cases of CAPA were diagnosed postmortem [61], and most patients with severe COVID-19 pneumonia are often too severely sick and hemodynamically unstable to undergo invasive diagnostic procedures such as bronchoscopy and lavage or lung biopsy. Furthermore, such procedures are contraindicated in COVID-19 patients due to the high risk of generating aerosols that are harmful to both patients and healthcare workers [62, 63].
Invasive pulmonary aspergillosis was diagnosed in 23% of critically ill patients with H1N1 viral infection, a median of 3 days after ICU admission, and corticosteroid use was found to be an independent risk factor for this superinfection [64]. Influenza was identified as an independent risk factor for patients diagnosed with invasive pulmonary aspergillosis and, thus, was associated with high mortality [65]. Previously, patients were often misdiagnosed with rhinocerebral aspergillosis, and invasive aspergillosis was also misdiagnosed, which was confused with a malignant disease [66]. Furthermore, image features of sino-orbital aspergillosis are nonspecific and may be confused with various orbital pathologies, and most common is an idiopathic orbital inflammatory disease [67, 68]. The involvement of the paranasal sinuses is usually a helpful clue in diagnosis, although it may not be present in certain cases; for example, concomitant sinus disease has been reported in 60–90% of cases in previous literature [69]. Orbital aspergillosis has also been misdiagnosed several times as malignancy [70, 71], optic neuritis [72], orbital apex syndrome [73], and typical bacterial cellulitis with orbital abscess [74].
Cross talk between clinical and environmental antifungal resistance Aspergillus fumigatus
The extensive application of fungicides containing active ingredients such as azoles and their interminable distribution in the environment are the link between clinical and environmental AFR strains of A. fumigatus [73, 74, 75••] (Fig. 1). In addition, recent studies have shown that AFR A. fumigatus strains in patients with invasive aspergillosis arise from environmental sources rather than through de novo mutation and selection in patients during antifungal drug treatment [24, 76, 77]. Several ecological hotspot niches contain AFR A. fumigatus strains, which includes flowerbeds, compost, leaves, seeds, soil, paddy fields, hospital environments, and hospital air samples [75, 78].
Possible routes of prevalence of antifungal-resistant Aspergillus fumigatus from the environment to the clinical health system through human inhalation. Application of chemical azole fungicides similar to medical azoles in an environment may result in the selection of azole-resistant A. fumigatus strains in the environment. Therefore, azole-susceptible individual may develop azole-resistant infections after inhaling these already azole-resistant strains. In the clinical healthcare system, azole-resistant A. fumigatus develops in immunocompromised individuals receiving long-term azole therapy for chronic aspergillosis. Figure created with http://biorender.com.
Clinically isolated strains have shown cross-resistance to voriconazole, posaconazole, itraconazole, and six triazole fungicides extensively used in agriculture, contributing to the widespread dissemination of multidrug-resistant pathogenic fungi in patients and hospitals [79]. While horizontal gene transfer is an important mechanism for the spread of antibiotic-resistant genes among fungal pathogens, conducting such a study is imperative because it has only been studied in bacteria. However, once multidrug-resistant genotypes merge in fungal pathogens, such genotypes can spread very quickly to other geographical regions and ecological niches through vegetative cells and airborne spores [80, 81]. Previous studies from several countries have shown that clinical strains of AFR A. fumigatus are associated with the use of the mentioned fungicides in agriculture and that these resistant strains have identical mutations of TR34/L98H and TR46/Y121F/T289A clones have been found worldwide from both environmental and clinical sources [81, 82]. AFR A. fumigatus with the TR34/L98H mutations was identified in the environment of Columbia [83], Portugal [84], Thailand [85], Netherlands [86], and the USA [77]. Clinically isolated strains have showed cross-resistance to voriconazole, posaconazole, itraconazole, and six triazole fungicides extensively used in agriculture, contributing to the widespread dissemination of multidrug-resistant pathogenic fungi in patients and hospitals [79].
Mode of action and mechanisms of resistance
The mode of action (MoA) of all major groups of antifungal agents such as azoles, echinocandins, and polyenes is similar, as they regularly influence cell structure and rigidity by interacting with cell wall or cell membrane components (Fig. 2A). Azoles are classified as either imidazoles or triazoles, which targets ergosterol biosynthetic pathway in fungal cells by inhibiting the cytochrome P450-dependent enzyme and lanosterol demethylase (LD), which is encoded by the Erg11 and Cyp51 genes [87, 88]. The LD enzyme plays an essential role in the synthesis of ergosterol; inhibition of the enzyme results in the accumulation of sterol precursors and 14α-methylated sterols in the fungal cell membranes [89, 90]. This means that azole exposure in the fungal cells leads to a reduction in ergosterol, altered plasma membrane structure, leakage of cell contents, increased cell permeability, and impaired growth [91, 92]. Therefore, it is important that azoles effectively bind to fungal lanosterol 14α-sterol-demethylase without affecting drug metabolism in the host. Echinocandins have a distinctive mode of action by inhibiting β-1-3-glucan synthase, which leads the breakdown of glucans that are important for cell wall components of several fungi [93]. Finally, polyenes have an unusual mode of action compared to other antifungal drug classes, as they do not bind to a specific enzyme, but instead interact with an important molecule—ergosterol, creating channels in the fungal cell membrane and killing cells by allowing ions and other cellular components to escape [94••].
Schematic representation showing the mode of actions of the primary antifungal drug classes in a fungal cell (A) and the known mechanisms of antifungal drug resistance in a fungal cell (B)—number (1) refers to mutations in Fks1/2 that inhibit echinocandins from binding and blocking the drug target 1,3-β-d-glucan synthase, (2) mutations in Erg11 that leads to overexpression of drug target lanosterol demethylase (LD) protein, (3) mutations in Cyp51A that inhibit azoles from binding and blocking LD, (4) increased efflux activity or transporter expression, e.g., ATP-binding cassette (ABC) or major facilitator superfamily (MFS) transporters, (5) ergosterol reduction, and (6) changes in ploidy. Figure created with PowerPoint in http://microsoftoffice.com.
Resistance mechanisms of AFR can be either intrinsic or acquired [95, 96]. Intrinsic resistance includes inherent resistance to antimicrobial drugs; for example, A. fumigatus [97] and other Aspergillus sp. [98] are intrinsically resistant to fluconazole. In contrast, acquired resistance is usually a result of exposure to antimicrobial selective pressures and can be caused by mutations, overexpression of resistance gene products, and/or genome rearrangements [99, 100••] (Fig. 2B). Furthermore, several resistance mechanisms of AFR in fungal strains may be because of upregulation of drug efflux and resistance genes or mutation [101], as the efflux pumps are transmembrane proteins that can efficiently transport drugs outside of the cell, thereby decreasing intracellular drug concentrations [102, 103]. In addition, overexpression due to upregulation of Erg11/Cyp51A genes that encode efflux transporters results in overexpression of these proteins [104]. Finally, mutations in the amino acid sequence of drug targets can also lead to resistance [105]. The most distinctive resistance mechanisms to AFR include the alterations in aneuploids and ergosterol synthesis. For instance, aneuploids are cells that have more or fewer chromosomes than the usual number [106, 107] and have recently been linked to azole AFR [108, 109]. The reduction of ergosterol synthesis through changes in expression or mutation has been shown to result in resistance to polyenes and amphotericin B [94].
Collaborative One Health approach for antifungal resistance Aspergillus fumigatus
A collaborative One Health approach that includes contributions at local, regional, national, and global levels could help manage AFR in Africa. Human, animal, and environmental health are closely linked, and responding to the threat of AFR requires the concerted efforts of all stakeholders such as mycologists (clinical, veterinary, and plant), farmers, and fungicide manufacturers as well as policymakers [110•].
Engaging employees from different sectors will change institutional and sociocultural beliefs to address this threat. Raising awareness through stakeholder networking will significantly reduce the risk and burden of AFR, thereby preserving the efficacy of AFR drugs like azole for clinical use [81]. The primary prevention and control measures for AFR include community engagement through media advertising (radio, television, or social media) and farmer meetings within the farm settlements. Therefore, education and dissemination of information on the effects of excessive and excessive use of azole fungicides on human, animal, and environmental health will be of significant benefit.
By bridging knowledge gaps about the impacts of fungicides and agricultural practices (composting), indiscriminate practices are harnessed. Information such as the effectiveness of applying low fungicide doses, aerating composting, rotating, or mixing azole fungicides with another fungicide with different modes of action limits resistance, reduces the development of azole resistance, and prolongs the benefits of azole [111, 112]. Education about the use of fungicides to prevent disease rather than promote plant growth is an alternative community orientation strategy.
Establishing antifungal stewardship in public health and the environment will improve and promote the appropriate use of azole and limit the risk of cross-resistance [113]. This is achieved by regulating the production and sale of fungicides and obtaining approval before bringing new fungicides to market. In addition, azole fungicides could be withdrawn from the market or replaced by another fungicide with a different mechanism of action or structure and fungicides with the same mode of action as medicinal azole will not be approved. Likewise, avoiding the use of fungicides in gardening or ornamental plant cultivation minimizes harmful effects and reduces the risk of resistance development. In addition, guidelines on dosage and frequency of fungicide application and breeding of resistant plants will limit the risk of cross-resistance and prevent dependence on fungicides [81].
In Africa, surveillance data on AFR is limited, and the prevalence of AFR is still underestimated. Therefore, collaboration between different sectors is urgently needed to enable informed decision-making, advocacy, and policymaking in rationalizing the available medical azole against azole resistance. Coordinated data sharing provides high-quality data to fill surveillance gaps. Creating an efficient standardized data collection registry requires the commitment of all stakeholders such as mycologists (clinical, animal, and plant), environmentalists, and government authorities. The data will increase knowledge and help determine the incidence of AFR across countries and regions. Therefore, addressing the extent of azole resistance in Africa cannot be emphasized due to the unavailability and inaccessibility of alternative therapies.
Interdisciplinary surveillance between researchers and stakeholders should be promoted. Multinational studies on AFR and discovering other potential resistance hotspots in Africa will generate data and solutions useful for public health interventions. In addition, the interlaboratory collection of Aspergillus strains will contribute to the knowledge of AFR incidence through collaboration with equipped clinical, local, regional, and reference laboratories with the capacity for susceptibility testing within a country or outside the African region. Regional data collection provides local data for informed planning and data-driven decisions within a community or place that are of value to a country. Finally, the lack of laboratory capacity to perform antifungal susceptibility testing (AFST) in most of our clinical laboratories requires collaboration with international organizations in capacity building to bridge this disparity.
Future perspective
The review studies described here have provided valuable insights into many aspects surrounding AFR A. fumigatus and collectively reported current approaches that could potentially mitigate the pathogen. However, the upsurge needs for improved treatment, and management of aspergillosis fungal pathogens is an ongoing challenge [6, 30, 114]. Special attention should be considered to applying new approaches such as drug repurposing, whole-genome sequencing, and RNA-based technology. Therefore, it is paramount to advance our understanding by deciphering the AFR A. fumigatus.
Drug repurposing
Development of repurposed drugs was essentially based on conventional drug delivery, consisting of de novo identification and new molecular units [115]. It is a time-consuming and an expensive process with a high risk of error [116, 117]. However, modern omics technology coupled with systems biology and high-throughput drug screenings enables a novel approach to drug repositioning in mitigating human diseases [118, 119]. Considering the repurposing of drugs to treat emerging diseases, this is a tremendous advantage when certain drugs are not yet available or developed. One of the major challenges in the global fight against infectious diseases is the inconsistent occurrence of diseases and the lack of treatment options for rare diseases, like the novel coronavirus disease (COVID-19) [120, 121] and currently aspergillosis in COVID-19 patients and individuals who have recovered from COVID-19 [122, 123].
The pathway to patient-specific personalization of drugs is important in the advancement and treatment of aspergillosis since the efficacy of drugs can be very dependent on different gene profiles because of the heterogeneity of human diseases [124, 125]. Human diseases are expressed through complex mechanisms that can be ascertained from several sources such as genetic aberrations, infectious diseases, and degenerative diseases [126]. In general, diseases often involve many intricate signaling cascades that vary greatly in specific individuals [127]. Individuals in different human populations have specific sets of inherited or non-inherited genetic abnormalities that can make certain individuals less responsive or unresponsive to common treatments or drugs [128]. In addition, registered drugs may be worthless to a particular individual if a specific drug target is missing and may not elicit the general response to a particular drug [129]. Therefore, it is crucial for drug personalization to reduce the lack of drug efficacy and drug repurposing.
Whole-genome sequencing
Establishing important analytical approaches to understand the acquisition of AFR A. fumigatus in Africa is fundamental to mitigating and controlling invasive fungal infections in patients and clinical environments. Regarding the advent of whole-genome sequencing (WGS) and its accessibility to the scientific domain, new diagnostic assessments using high-throughput DNA sequencing of next-generation sequencing (NGS) approach can provide a significant alternative for diagnostics and establish knowledge of fungal biodiversity for epidemiological purposes [114, 130]. This technique has been revolutionized and is currently being used to study the association between various fungal strains isolated from immunocompromised patients when a cluster of cases occurs in order to comprehend disease outbreaks. It provides large sample sizes, allows evaluation of mutation frequencies that correlate with resistance, and identifies the most dominant community. For example, the successful studies on hospital outbreaks of aspergillosis in immunocompromised patients [131] and in transplant patients were attained using NGS analysis [132, 133•]. In addition, studying the correlation of the sequence of genes associated with resistance in fungal and clinical environmental isolates showing resistance requires understanding how the mutation may affect cell composition linked to drug susceptibility [100]. Another complementary approach is to examine the mechanisms of antifungal resistance with less susceptible strains and higher minimal inhibitory concentration (MIC) to a drug, compared to the control [134].
RNA-based technology
The last decade has produced led to an increase in the number of molecular techniques available to understand the function of genes involved in pathogenesis. However, RNA interference (RNAi) and clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein (Cas-9) (CRISPR-Cas9)-based technology are the leading approaches that can provide valuable insights into aspergillosis [135, 136], its associated pathogens and, thus, have the potential to uncover new and interesting biological and clinical information.
RNA interference pathways are highly conserved in eukaryotes to negatively regulate gene expression by short RNAs or small non-coding RNAs (sRNAs) [137]. The established RNAi pathway produces double-stranded RNA (dsRNA) by RNA-dependent RNA polymerases, which are eventually processed by Dicer enzymes to produce the sRNAs (Fig. 3). Thus, these endogenous RNAs are used to suppress different target sequences [138, 139•]. RNAi is initiated by introducing a dsRNA, which is homologous to the target sequence. The dsRNA is processed to 21–25 nucleotides by an endonuclease known as DICER. One strand of the resulting small interfering RNAs (siRNA) associates with the effector protein, Argonaute, which then binds and degrades target mRNA in a homology-dependent manner, silencing gene transcript levels.
Schematic representation of RNAi-mediated silencing mechanism in fungi. Figure created with PowerPoint in Microsoft http://office.com.
CRISPR/Cas-9 mechanism generates specific double-stranded pieces at the target locus that trigger DNA repairs [140•] (Fig. 4). The designed single guide RNA (sgRNA) identifies the target sequence in the gene of interest through a complementary base pair. While the Cas-9 nuclease creates double-stranded pieces at a location, three base pairs upstream of the protospacer adjacent motif, then the double-stranded piece becomes cellular repaired either by knockouts (KO) through non-homologous end joining or knock-ins (KI) by homologous recombination. Until date, the Aspergillus genus of the A. fumigatus offers the largest range of molecular genetic tools for studying gene function [141, 142]. Other related aspergillosis-causing genera lack similar genetic toolkits, and only the RNAi technology was accessible in A. oryzae [143]. Fortunately, this grim situation is changing owing to the extension of CRISPR-Cas9 technological approach against relevant medical fungi. However, the methods should be optimized as they only work in genes that generate a selectable phenotype upon mutation, making their widespread use impossible. In light of this, a promising plasmid-free CRISPR-Cas9-based method was established that produces stable transformants in Candida [144], Cryptococcus [145], and Aspergillus sp. [146] which can facilitate a rapid process by allowing the targeted mutation of any gene and the use of microhomology repair templates [147].
Schematic representation of CRISPR/Cas9-mediated gene editing mechanism in fungi. Figure created with http://biorender.com.
Special attention should also be given to deciphering the mechanisms that confer intrinsic and acquired resistance to antifungal drugs, in particular which genes of Aspergillus fungi are expressed during aspergillosis. It is worth noting that the existing circumstances hamper the progress of treatments to tackle the disease, as advances made in one species are not confirmed in detail in other species or genera. This vital question has been very challenging to answer for technical reasons. Precisely, when isolating total RNA from a host that has been infected with any microbe, thus, the signal from host transcripts usually overcomes the signal from the infecting microbe, and the pathogen RNA consists of only a tiny fraction, about 0.1% of the total amount of RNA extracted [148, 149]. The successful improvement approaches to screen for an enrich fungal transcript from total RNA samples obtained from infected mouse tissues have been applied in vivo in mouse infection models of A. fumigatus [150, 151]. Therefore, applying careful enrichment RNA transcript methods to investigate the expression of AFR genes on variable isolates of A. fumigatus in mouse models of aspergillosis could certainly elucidate important virulence genes that could potentially be useful as diagnostic biomarkers.
The current innovative approaches to understanding and mitigating the function of genes involved in pathogenesis are RNAi and CRISPR-Cas9-based technology [136]. Thus, these approaches have the potential to uncover new and interesting biological and clinical information of invasive fungal pathogens. Nonetheless, it is crucial that we use comprehensive research tools to combat antifungal pathogens. Therefore, it is of crucial importance to use such techniques in the mitigation and control of AFR A. fumigatus in Africa, thereby improving the treatment of aspergillosis disease.
Conclusions
The AFR A. fumigatus is widely recognized as a threat to humans throughout the world and the risk posed by this emerging pathogen across Africa, as well as the level of antifungal exposure in the environment and its impact on AFR in humans are understudied. Given the incomplete removal or inactivation of antifungal agents during agricultural practices and the direct application of effective antifungal concentrations to agricultural products, the environmental dimension of AFR requires greater attention since commercial and subsistence agriculture is generally practiced in African countries. The potential and expectation of insight into the recent advances and future perspective strategies can improve the effectiveness of emerging antifungal-resistant pathogen in Africa, and thus, it is critical to invest in reducing the burden and enhancing the clinical outcome of diseases.
To combat AFR A. fumigatus in Africa, it is important to improve current diagnostic tools (e.g., antifungal drug susceptibility) and the availability of these tests in poor and disadvantaged settings. In addition, the use of next-generation and metagenomic sequencing has the potential to enable screening of this pathogen. Community engagement and advocacy to improve access to safe use of azole fungicides and effective therapy with triazoles in clinical settings are needed. However, a better understanding of the resistance mechanisms of this fungal pathogen is promising for the development of new strategies to target this pathogen while strengthening the host immune response. Comprehending this resistance mechanism will facilitate the high-throughput screening applications, with the CRISPR/Cas9 system and RNAi silencing–mediated approach are at the forefront of this development on inhibiting known genes associated with AFR resistance. Ultimately, if these novel high-throughput approaches are used to combat the invasive AFR pathogen A. fumigatus in Africa, it can be expected that further improvements in this technology will be achieved, and some of the challenges such as efficiency will be overcome in the future.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Friedman DZP, Schwartz IS. Emerging fungal infections: new patients, new patterns, and new pathogens. J Fungi. 2019;5:67.
Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4
Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases - estimate precision. J Fungi. 2017:3.
Samrah S, Sweidan A, Aleshawi A, Ayesh M. Fusarium-induced cellulitis in an immunocompetent patient with sickle cell disease: a case report. J Investig Med High Impact Case Rep. 2020:8.
Bellanger AP, Navellou JC, Lepiller Q, Brion A, Brunel AS, Millon L, et al. Mixed mold infection with Aspergillus fumigatus and Rhizopus microsporus in a severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) patient. Infectious Diseases Now. 2021;51:633.
Truda VSS, Falci DR, Porfírio FMV, de Santos DW Junior FI, Pasqualotto AC, et al. A contemporary investigation of burden and natural history of aspergillosis in people living with HIV/AIDS. Mycoses 2023
Song G, Liang G, Liu W. Fungal co-infections associated with global COVID-19 pandemic: a clinical and diagnostic perspective from China. Mycopathologia. 2020;185:599–606.
Bhatt K, Agolli A, Patel MH, Garimella R, Devi M, Garcia E, et al. High mortality co-infections of COVID-19 patients: mucormycosis and other fungal infections. Discoveries. 2021;9:e126.
Rodrigues ML, Nosanchuk JD. Fungal diseases as neglected pathogens: a wake-up call to public health officials. PLoS Negl Trop Dis. 2020;14
WHO. First meeting of the WHO antifungal expert group on identifying priority fungal pathogens. 2020.
WHO. WHO fungal priority pathogens list to guide research development and public health. World Health Organization; 2022. p. 1–48. An outstanding document that systematically prioritizes fungal pathogens with the goal of guiding future research and policy changes to improve the global response to fungal infections and antifungal resistance.
Driemeyer C, Falci DR, Oladele RO, Bongomin F, Ocansey BK, Govender NP, et al. The current state of clinical mycology in Africa: a European Confederation of Medical Mycology and International Society for Human and Animal Mycology survey. The Lancet Microbe. 2022;
WHO. WHO guidelines for the diagnosis, prevention and management of cryptococcal disease; 2018. p. 1–62.
Nasir I, Shuwa H, Emeribe A, Adekola H, Dangana A. Phenotypic profile of pulmonary aspergillosis and associated cellular immunity among people living with human immunodeficiency virus in Maiduguri, Nigeria. Tzu-Chi Medical Journal. 2019;31:153.
Wiederhold NP. Antifungal resistance: current trends and future strategies to combat. Infect Drug Resist. 2017;10:259.
Wiederhold NP. Emerging fungal infections: new species, new names, and antifungal resistance. Clin Chem. 2022;68:90.
Campbell CA, Osaigbovo II, Oladele RO. Triazole susceptibility of Aspergillus species: environmental survey in Lagos, Nigeria and review of the rest of Africa. Ther Adv Infect Dis. 2021;8:20499361211044330.
Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 2018;360:739–42.
Hendrickson JA, Hu C, Aitken SL, Beyda N. Antifungal resistance: a concerning trend for the present and future. Curr Infect Dis Rep. 2019;21:1–8.
Rodrigues CF, Henriques M. Oral mucositis caused by Candida glabrata biofilms: failure of the concomitant use of fluconazole and ascorbic acid. Ther Adv Infect Dis. 2017;4:10–7.
Bhattacharya S, Sae-Tia S, Fries BC. Candidiasis and mechanisms of antifungal resistance. Antibiotics. 2020;9:312.
Rybak JM, Cuomo CA, David RP. The molecular and genetic basis of antifungal resistance in the emerging fungal pathogen Candida auris. Curr Opin Microbiol. 2022;70:102208.
Wang Q, Lei C, Cheng H, Yang X, Huang Z, Chen X, et al. Widespread dissemination of plasmid-mediated tigecycline resistance gene tet (X4) in Enterobacterales of porcine origin. Microbiol Spectr. 2022;10:e01615–22.
Rhodes J, Abdolrasouli A, Dunne K, Sewell TR, Zhang Y, Ballard E, et al. Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment. Nat Microbiol. 2022;7:663–74.
O’brien B, Chaturvedi S, Chaturvedi V. In vitro evaluation of antifungal drug combinations against multidrug-resistant Candida auris isolates from New York outbreak. Antimicrob Agents Chemother. 2020;64:e02195–19.
Oladele R, Uwanibe JN, Olawoye IB, Ettu AWO, Meis JF, Happi CT. Emergence and genomic characterization of multidrug resistant Candida auris in Nigeria, West Africa. J Fungi. 2022;8:787. The authors of this study used whole-genome sequencing (WGS) to identify the emerging multidrug-resistant fungal pathogens. The study is important for public health because it highlights the need for active disease surveillance in Africa as well as diagnostic gaps in healthcare settings.
Lestrade PP, Bentvelsen RG, Schauwvlieghe AFAD, Schalekamp S, Van Der Velden WJFM, Kuiper EJ, et al. Voriconazole resistance and mortality in invasive aspergillosis: a multicenter retrospective cohort study. Clin Infect Dis. 2019;68:1463–71.
Chen J, Li H, Li R, Bu D, Wan Z. Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J Antimicrob Chemother. 2005;55:31–7.
Chen P, Liu M, Zeng Q, Zhang Z, Liu W, Sang H, et al. Uncovering new mutations conferring azole resistance in the Aspergillus fumigatus cyp51A gene. Front Microbiol. 2020;10:3127–38.
Salehi Z, Sharifynia S, Jamzivar F, Shams-Ghahfarokhi M, Poorabdollah M, Abtahian Z, et al. Clinical epidemiology of pulmonary aspergillosis in hospitalized patients and contribution of Cyp51A, Yap1, and Cdr1B mutations to voriconazole resistance in etiologic Aspergillus species. Eur J Clin Microbiol Infect Dis. 2023;2023:1–12.
Fang W, Wu J, Cheng M, Zhu X, Du M, Chen C, et al. Diagnosis of invasive fungal infections: challenges and recent developments. J Biomed Sci. 2023;30:42.
Fosses Vuong M, Hollingshead CM, Waymack JR. Aspergillosis. Treasure Island (FL): StatPearls; 2023. http://www.ncbi.nlm.nih.gov/books/NBK482241/. Accessed 11 Sep 2023
Danion F, Rouzaud C, Duréault A, Poirée S, Bougnoux M-E, Alanio A, et al. Why are so many cases of invasive aspergillosis missed? Med Mycol. 2019;57:S94–103.
Rasheed W, Tasnim S, Dweik A, Al-Jabory O, Usala S. Allergic bronchopulmonary aspergillosis (ABPA) diagnosis missed in the context of asthma exacerbation due to medication nonadherence. Cureus. 2022;14:e28202.
Denham ST, Wambaugh MA, Brown JCS. How environmental fungi cause a range of clinical outcomes in susceptible hosts. J Mol Biol. 2019;431:2982–3009.
Banerjee S, Denning DW, Chakrabarti A. One Health aspects & priority roadmap for fungal diseases : a mini-review. Indian J Med Res. 2021;153:311–9.
Guarner J, Brandt ME. Histopathologic diagnosis of fungal infections in the 21st century. Clin Microbiol Rev. 2011;24:247–80.
Sangoi AR, Rogers WM, Longacre TA, Montoya JG, Baron EJ, Banaei N. Challenges and pitfalls of morphologic identification of fungal infections in histologic and cytologic specimens: a ten-year retrospective review at a single institution. Am J Clin Pathol. 2009;131:364–75.
Lee S, Yun NR, Kim K-H, Jeon JH, Kim E-C, Chung DH, et al. Discrepancy between histology and culture in filamentous fungal infections. Med Mycol. 2010;48:886–8.
Shah AA, Hazen KC. Diagnostic Accuracy of Histopathologic and cytopathologic examination of aspergillus species. Am J Clin Pathol. 2013;139:55–61.
Arvanitis M, Anagnostou T, Fuchs BB, Caliendo AM, Mylonakis E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin Microbiol Rev. 2014;27:490–526.
Bassetti M, Peghin M, Vena A. Challenges and solution of invasive aspergillosis in non-neutropenic patients: a review. Infect Dis Ther. 2018;7:17–27.
Russo A, Tiseo G, Falcone M, Menichetti F. Pulmonary aspergillosis: an evolving challenge for diagnosis and treatment. Infect Dis Ther. 2020;9:511–24.
Negroni R. Onychomycosis due to aspergillus species. In: Comarú Pasqualotto A, editor. Aspergillosis: From Diagnosis to Prevention. Dordrecht: Springer Netherlands; 2010. p. 961–71. https://doi.org/10.1007/978-90-481-2408-4_56.
Bongomin F, Batac CR, Richardson MD, Denning DW. A review of onychomycosis due to aspergillus species. Mycopathologia. 2018;183:485–93.
Chakrabarti A, Chatterjee S, Radotra BD. Cutaneous and wound aspergillosis. In: Comarú Pasqualotto A, editor. Aspergillosis: From Diagnosis to Prevention. Dordrecht: Springer Netherlands; 2010. p. 939–59. https://doi.org/10.1007/978-90-481-2408-4_55.
Bongomin F, Asio LG, Baluku JB, Kwizera R, Denning DW. Chronic pulmonary aspergillosis: notes for a clinician in a resource-limited setting where there is no mycologist. J Fungi (Basel). 2020;6:75. An excellent review that highlights the crucial knowledge needed by a non-specialist mycologist or clinician in a resource-constrained setting to diagnose and treat chronic pulmonary aspergillosis (CPA), which is brought on by Aspergillus species in immunocompromised patients.
Denning DW, Pfavayi LT. Poorly controlled asthma – easy wins and future prospects for addressing fungal allergy. Allergol Int. 2023;72:493–506.
Hughes KM, Price D, Torriero AAJ, Symonds MRE, Suphioglu C. Impact of fungal spores on asthma prevalence and hospitalization. Int J Mol Sci. 2022;23:4313.
Rodrigues ML, Albuquerque PC. Searching for a change: the need for increased support for public health and research on fungal diseases. PLoS Negl Trop Dis. 2018;12:e0006479.
Zhang R, Wang S, Lu H, Wang Z, Xu X. Misdiagnosis of invasive pulmonary aspergillosis: a clinical analysis of 26 immunocompetent patients. Int J Clin Exp Med. 2014;7:5075–82.
Setianingrum F, Rozaliyani A, Adawiyah R, Syam R, Tugiran M, Sari CYI, et al. A prospective longitudinal study of chronic pulmonary aspergillosis in pulmonary tuberculosis in Indonesia (APICAL). Thorax. 2022;77:821–8.
Agarwal R, Chakrabarti A. Clinical manifestations and natural history of allergic bronchopulmonary aspergillosis. In: Comarú Pasqualotto A, editor. Aspergillosis: From Diagnosis to Prevention. Dordrecht: Springer Netherlands; 2010. p. 707–24. https://doi.org/10.1007/978-90-481-2408-4_42.
Lai C-C, Wang C-Y, Hsueh P-R. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J Microbiol Immunol Infect. 2020;53:505–12.
Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762–8.
Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–74.
Lai C-C, Yu W-L. Appropriate use of antimicrobial therapy for COVID-19 co-infection. Immunotherapy. 2021; https://doi.org/10.2217/imt-2021-0134.
Janssen NAF, Nyga R, Vanderbeke L, Jacobs C, Ergün M, Buil JB, et al. Multinational observational cohort study of COVID-19-associated pulmonary aspergillosis1. Emerg Infect Dis. 2021;27:2892–8.
Verweij PE, Gangneux J-P, Bassetti M, Brüggemann RJM, Cornely OA, Koehler P, et al. Diagnosing COVID-19-associated pulmonary aspergillosis. The Lancet Microbe. 2020;1:e53–5.
Bartoletti M, Pascale R, Cricca M, Rinaldi M, Maccaro A, Bussini L, et al. Epidemiology of invasive pulmonary aspergillosis among intubated patients with COVID-19: a prospective study. Clin Infect Dis. 2021;73:e3606–14.
Mohamed A, Rogers TR, Talento AF. COVID-19 Associated invasive pulmonary aspergillosis: diagnostic and therapeutic challenges. J Fungi (Basel). 2020;6:115.
Wahidi MM, Lamb C, Murgu S, Musani A, Shojaee S, Sachdeva A, et al. American association for bronchology and interventional pulmonology (AABIP) statement on the use of bronchoscopy and respiratory specimen collection in patients with suspected or confirmed COVID-19 infection. J Bronchology Interv Pulmonol. 2020;27:e52–4.
Wahidi MM, Shojaee S, Lamb CR, Ost D, Maldonado F, Eapen G, et al. The use of bronchoscopy during the Coronavirus disease 2019 pandemic: CHEST/AABIP guideline and expert panel report. Chest. 2020;158:1268–81.
Wauters J, Baar I, Meersseman P, Meersseman W, Dams K, De Paep R, et al. Invasive pulmonary aspergillosis is a frequent complication of critically ill H1N1 patients: a retrospective study. Intensive Care Med. 2012;38:1761–8.
Schauwvlieghe AFAD, Rijnders BJA, Philips N, Verwijs R, Vanderbeke L, Van Tienen C, et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir Med. 2018;6:782–92.
Chang T, Teng MMH, Wang SF, Li WY, Cheng CC, Lirng JF. Aspergillosis of the paranasal sinuses. Neuroradiology. 1992;34:520–3.
Austin P, Dekker A, Kennerdell JS, Orbital aspergillosis. Report of a case diagnosed by fine needle aspiration biopsy. Acta Cytol. 1983;27:166–9.
Adulkar NG, Radhakrishnan S, Vidhya N, Kim U. Invasive sino-orbital fungal infections in immunocompetent patients: a clinico-pathological study. Eye (Lond). 2019;33:988–94.
Heier JS, Gardner TA, Hawes MJ, McGuire KA, Walton WT, Stock J. Proptosis as the initial presentation of fungal sinusitis in immunocompetent patients. Ophthalmology. 1995;102:713–7.
Mauriello JA, Yepez N, Mostafavi R, Barofsky J, Kapila R, Baredes S, et al. Invasive rhinosino-orbital aspergillosis with precipitous visual loss. Can J Ophthalmol. 1995;30:124–30.
Cullen GD, Davidson TM, Yetmar ZA, Pritt BS, DeSimone DC. Orbital apex syndrome due to invasive aspergillosis in an immunocompetent patient. IDCases. 2021;25:e01232.
Mori S, Kurimoto T, Kawara K, Ueda K, Sakamoto M, Keshi Y, et al. The difficulty of diagnosing invasive aspergillosis initially manifesting as optic neuropathy. Case Reports in Ophthalmology. 2019;10:11–8.
Chang Y-M, Chang Y-H, Chien K-H, Liang C-M, Tai M-C, Nieh S, et al. Orbital apex syndrome secondary to aspergilloma masquerading as a paranasal sinus tumor. Medicine (Baltimore). 2018;97:e11650.
Marcet MM, Yang W, Albert DM, Salamat MS, Appen RE. Aspergillus infection of the orbital apex masquerading as Tolosa-Hunt syndrome. Arch Ophthalmol. 2007;125:563–6.
Burks C, Darby A, Londoño LG, Momany M, Brewer MT. Azole-resistant Aspergillus fumigatus in the environment: identifying key reservoirs and hotspots of antifungal resistance. PLoS Pathog. 2021;17:e1009711. An excellent review highlighting the mutations of azole-resistant Aspergillus fumigatus that emerged in agricultural settings and the link on the world map showing where resistant isolates have been found between environmental and clinical settings.
Barber AE, Riedel J, Sae-Ong T, Kang K, Brabetz W, Panagiotou G, et al. Effects of agricultural fungicide use on Aspergillus fumigatus abundance, antifungal susceptibility, and population structure. mBio. 2020;11 https://doi.org/10.1128/mbio.02213-20.
Bradley K, Le-Mahajan A, Morris B, Peritz T, Chiller T, Forsberg K, et al. Fatal fungicide-associated triazole-resistant Aspergillus fumigatus infection, Pennsylvania, USA. Emerg Infect Dis. 2022;28:1904–5.
van der Torre MH, Whitby C, Eades CP, Moore CB, Novak-Frazer L, Richardson MD, et al. Absence of azole antifungal resistance in Aspergillus fumigatus isolated from root vegetables harvested from UK arable and horticultural soils. Journal of Fungi. 2020;6:208.
Resendiz-Sharpe A, Dewaele K, Merckx R, Bustamante B, Vega-Gomez MC, Rolon M, et al. Triazole-resistance in environmental Aspergillus fumigatus in Latin American and African countries. J Fungi (Basel). 2021;7:292.
Langfeldt A, Gold JAW, Chiller T. Emerging fungal infections: from the fields to the clinic, resistant Aspergillus fumigatus and dermatophyte species: a One Health perspective on an urgent public health problem. Curr Clin Micro Rpt. 2022;9:46–51.
Verweij PE, Lucas JA, Arendrup MC, Bowyer P, Brinkmann AJF, Denning DW, et al. The One Health problem of azole resistance in Aspergillus fumigatus: current insights and future research agenda. Fungal Biol Rev. 2020;34:202–14.
Sewell TR, Zhu J, Rhodes J, Hagen F, Meis JF, Fisher MC, et al. Nonrandom distribution of azole resistance across the global population of Aspergillus fumigatus. mBio. 2019;10:e00392-19.
Alvarez-Moreno C, Lavergne R-A, Hagen F, Morio F, Meis JF, Le Pape P. Azole-resistant Aspergillus fumigatus harboring TR34/L98H, TR46/Y121F/T289A and TR53 mutations related to flower fields in Colombia. Sci Rep. 2017;7:45631.
Gonçalves P, Melo A, Dias M, Almeida B, Caetano LA, Veríssimo C, et al. Azole-resistant Aspergillus fumigatus harboring the TR34/L98H mutation: first report in Portugal in environmental samples. Microorganisms. 2021;9:57.
Tangwattanachuleeporn M, Minarin N, Saichan S, Sermsri P, Mitkornburee R, Groß U, et al. Prevalence of azole-resistant Aspergillus fumigatus in the environment of Thailand. Med Mycol. 2017;55:429–35.
van Paassen J, Russcher A, In’t Veld-van Wingerden AW, Verweij PE, Kuijper EJ. Emerging aspergillosis by azole-resistant Aspergillus fumigatus at an intensive care unit in the Netherlands, 2010 to 2013. Euro Surveill. 2016:21.
Zhang J, Li L, Lv Q, Yan L, Wang Y, Jiang Y. The fungal CYP51s: their functions, structures, related drug resistance, and inhibitors. Front Microbiol. 2019;10:691–707.
Rosam K, Monk BC, Lackner M. Sterol 14α-demethylase ligand-binding pocket-mediated acquired and intrinsic azole resistance in fungal pathogens. Journal of Fungi. 2021;7:1.
Song J, Liu X, Li R. Sphingolipids: regulators of azole drug resistance and fungal pathogenicity. Mol Microbiol. 2020;114:891–905.
Osset-Trénor P, Pascual-Ahuir A, Proft M. Fungal drug response and antimicrobial resistance. J Fungi (Basel). 2023;9:565.
Bhattacharya S, Esquivel BD, White TC. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio. 2018;9:e01291-18.
Zangl I, Beyer R, Gattesco A, Labuda R, Pap I-J, Strauss J, et al. Limosilactobacillus fermentum limits Candida glabrata growth by ergosterol depletion. Microbiology Spectrum. 2023;11:e03326–2.
Szymański M, Chmielewska S, Czyżewska U, Malinowska M, Tylicki A. Echinocandins – structure, mechanism of action and use in antifungal therapy. J Enzyme Inhib Med Chem. 2022;37:876–94.
Carolus H, Pierson S, Lagrou K, Van Dijck P. Amphotericin B and other polyenes—discovery, clinical use, mode of action and drug resistance. J Fungi (Basel). 2020;6:321. An excellent review explaining the mode of actions of the primary antifungal drug classes in a fungal cell.
Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, et al. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol. 2022;20:557–71.
Hossain CM, Ryan LK, Gera M, Choudhuri S, Lyle N, Ali KA, et al. Antifungals and drug resistance. Encyclopedia. 2022;2:1722–37.
Leonardelli F, Macedo D, Dudiuk C, Cabeza MS, Gamarra S, Garcia-Effron G. Aspergillus fumigatus intrinsic fluconazole resistance is due to the naturally occurring T301I substitution in Cyp51Ap. Antimicrob Agents Chemother. 2016;60:5420–6.
Macedo D, Leonardelli F, Gamarra S, Garcia-Effron G. Emergence of triazole resistance in Aspergillus spp. in Latin America. Curr Fungal Infect Rep. 2021;15:93–103.
Berman J, Krysan DJ. Drug resistance and tolerance in fungi. Nat Rev Microbiol. 2020;18:319–31.
Lee Y, Robbins N, Cowen LE. Molecular mechanisms governing antifungal drug resistance. npj Antimicrob Resist. 2023;1:1–9. An excellent review explaining the mechanisms of antifungal drug resistance in a fungal cell.
Franconi I, Rizzato C, Poma N, Tavanti A, Lupetti A. Candida parapsilosis sensu stricto antifungal resistance mechanisms and associated epidemiology. Journal of Fungi. 2023;9:798.
Song J, Zhou J, Zhang L, Li R. Mitochondria-mediated azole drug resistance and fungal pathogenicity: opportunities for therapeutic development. Microorganisms. 2020;8:1574.
Nishino K, Yamasaki S, Nakashima R, Zwama M, Hayashi-Nishino M. Function and inhibitory mechanisms of multidrug efflux pumps. Frontiers in Microbiology. 2021;12 https://doi.org/10.3389/fmicb.2021.737288.
Handelman M, Osherov N. Experimental and in-host evolution of triazole resistance in human pathogenic fungi. Front Fungal Biol. 2022;3 https://doi.org/10.3389/ffunb.2022.957577.
Kuplińska A, Rząd K. Molecular targets for antifungals in amino acid and protein biosynthetic pathways. Amino Acids. 2021;53:991.
Gilchrist C, Stelkens R. Aneuploidy in yeast: segregation error or adaptation mechanism? Yeast. 2019;36:525–39.
Tosh J, Tybulewicz V, Fisher EMC. Mouse models of aneuploidy to understand chromosome disorders. Mamm Genome. 2022;33:157–68.
Sun L, Li H, Yan T, Cao Y, Jiang Y, Yang F. Aneuploidy enables cross-tolerance to unrelated antifungal drugs in Candida parapsilosis. Front Microbiol. 2023;14 https://doi.org/10.3389/fmicb.2023.1137083.
Yang F, Gritsenko V, Lu H, Zhen C, Gao L, Berman J, et al. Adaptation to fluconazole via aneuploidy enables cross-adaptation to amphotericin B and flucytosine in Cryptococcus neoformans. Microbiol Spectr. 2021;9:e00723-21.
Otu A, Osaigbovo I, Orefuwa E, Ebenso B, Ojumu T. Collaborative One Health approaches can mitigate increasing azole-resistant Aspergillus fumigatus in Africa. The Lancet Microbe. 2021;2:e490–1. An excellent review highlighting the importance of collaborative One Health approaches to improve and manage the prevalence of azole-resistant Aspergillus fumigatus in Africa.
Meyer M, Diehl D, Schaumann GE, Muñoz K. Agricultural mulching and fungicides—impacts on fungal biomass, mycotoxin occurrence, and soil organic matter decomposition. Environ Sci Pollut Res. 2021;28:36535–50.
Gikas GD, Parlakidis P, Mavropoulos T, Vryzas Z. Particularities of fungicides and factors affecting their fate and removal efficacy: a review. Sustainability. 2022;14:4056.
Woods M, McAlister JA, Geddes-McAlister J. A One Health approach to overcoming fungal disease and antifungal resistance. WIREs Mechanisms of Disease. 2023;15:e1610.
Bao S, Song H, Chen Y, Zhong C, Tang H. Metagenomic next-generation sequencing for the diagnosis of pulmonary aspergillosis in non-neutropenic patients: a retrospective study. Front Cell Infect Microbiol. 2022;12:925982.
Rudrapal M, Khairnar SJ, Jadhav AG. Drug repurposing (DR): an emerging approach in drug discovery. Drug Repurposing - Hypothesis. Mol Aspects and Ther Appl. 2020;
Xue H, Li J, Xie H, Wang Y. Review of drug repositioning approaches and resources. Int J Biol Sci. 2018;14:1232.
Pillaiyar T, Meenakshisundaram S, Manickam M, Sankaranarayanan M. A medicinal chemistry perspective of drug repositioning: recent advances and challenges in drug discovery. Eur J Med Chem. 2020;195:112275.
Kiriiri GK, Njogu PM, Mwangi AN. Exploring different approaches to improve the success of drug discovery and development projects: a review. Future J Pharm Sci. 2020;6:27.
Pandita V, Parihar A, Parihar DS, Panda S, Shanmugarajan D, Kumari L, et al. System and network biology-based computational approaches for drug repositioning. Computational Approaches for Novel Therapeutic and Diagnostic Designing to Mitigate SARS-CoV-2 Infection. 2022;267–290.
Low ZY, Farouk IA, Lal SK. Drug repositioning: new approaches and future prospects for life-debilitating diseases and the COVID-19 pandemic outbreak. Viruses. 2020:12.
Sultana J, Crisafulli S, Gabbay F, Lynn E, Shakir S, Trifirò G. Challenges for drug repurposing in the COVID-19 pandemic era. Front Pharmacol. 2020;11
Khodavaisy S, Khajavirad N, Hashemi SJ, Izadi A, Dehghan Manshadi SA, Abdollahi A, et al. Proven pulmonary aspergillosis in a COVID-19 patient: a case report. Curr Med Mycol. 2021;7:39–42.
Bhandari S, Gupta S, Bhargava S, Samdani S, Singh SN, Sharma BB, et al. COVID associated invasive aspergillosis. Indian J Otolaryngol Head Neck Surg. 2023;75:557–62.
Arastehfar A, Carvalho A, Houbraken J, Lombardi L, Garcia-Rubio R, Jenks JD, et al. Aspergillus fumigatus and aspergillosis: from basics to clinics. Stud Mycol. 2021;100:100115.
Verburg K, van Neer J, Duca M, de Cock H. Novel Treatment approach for aspergilloses by targeting germination. J Fungi (Basel). 2022;8:758.
Li J, Zhang Y, Li J, Sun T, Tian C. Metabolic engineering of the cellulolytic thermophilic fungus Myceliophthora thermophila to produce ethanol from cellobiose. Biotechnol Biofuels. 2020;13:1–15.
Valls PO, Esposito A. Signalling dynamics, cell decisions, and homeostatic control in health and disease. Curr Opin Cell Biol. 2022;75:102066.
Jackson M, Marks L, May GHW, Wilson JB. The genetic basis of disease. Essays Biochem. 2018;62:723.
Adepu S, Ramakrishna S. Controlled drug delivery systems: current status and future directions. Molecules. 2021;26:5905.
Reslan L, Araj GF, Finianos M, El Asmar R, Hrabak J, Dbaibo G, et al. Molecular characterization of Candida auris isolates at a major tertiary care center in Lebanon. Front Microbiol. 2022;12:3916.
Sung AH, Kiely G, Singh JK, Thomas S, Lough J, Smith M. Hospital-treated serious and invasive aspergillosis and candidiasis infections during the COVID-19 pandemic: a retrospective analysis of Hospital Episode Statistics data from England. BMJ Open. 2023;13:e070537.
Chen F, Zhao Y, Shen C, Han L, Chen X, Zhang J, et al. Next generation sequencing for diagnosis of central nervous system aspergillosis in liver transplant recipients. Ann Transl Med. 2021;9:1071.
Yu J, Diaz JD, Goldstein SC, Patel RD, Varela JC, Reyenga C, et al. Impact of next-generation sequencing cell-free pathogen DNA test on antimicrobial management in adults with hematological malignancies and transplant recipients with suspected infections. Transplantation Cell Ther. 2021;27:500.e1–6. In this study, the authors showed the in-depth information on the value of next-generation sequencing (NGS) for diagnosing aspergillosis in immunocompromised patients.
Pellaton N, Sanglard D, Lamoth F, Coste AT. How yeast antifungal resistance gene analysis is essential to validate antifungal susceptibility testing systems. Front Cell Infect Microbiol. 2022;12 https://doi.org/10.3389/fcimb.2022.859439.
Jiang C, Lv G, Tu Y, Cheng X, Duan Y, Zeng B, et al. Applications of CRISPR/Cas9 in the synthesis of secondary metabolites in filamentous fungi. Front Microbiol. 2021;12:182.
Bruch A, Kelani AA, Blango MG. RNA-based therapeutics to treat human fungal infections. Trends Microbiol. 2022;30:411–20.
Billmyre RB, Calo S, Feretzaki M, Wang X, Heitman J. RNAi function, diversity, and loss in the fungal kingdom. Chromosome Res. 2013;21:561–72.
Hoehener C, Hug I, Nowacki M. Dicer-like enzymes with sequence cleavage preferences. Cell. 2018;173:234–247.e7.
Di Fazio A, Schlackow M, Pong SK, Alagia A, Gullerova M. Dicer dependent tRNA derived small RNAs promote nascent RNA silencing. Nucleic Acids Res. 2022;50:1734–52. An excellent review highlighting the pathways for RNA interference (RNAi)–mediated silencing mechanism in fungi.
Asmamaw M, Zawdie B. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics. 2021;15:353–61. An excellent review describing the CRISPR/Cas9-mediated gene editing mechanism in fungi.
de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017;18:28.
Gupta SK, Srivastava M, Osmanoglu Ö, Xu Z, Brakhage AA, Dandekar T. Aspergillus fumigatus versus genus Aspergillus: conservation, adaptive evolution and specific virulence genes. Microorganisms. 2021;9:2014.
Yamada O, Ikeda R, Ohkita Y, Hayashi R, Sakamoto K, Akita O. Gene silencing by RNA interference in the koji mold Aspergillus oryzae. Biosci Biotechnol Biochem. 2007;71:138–44.
Maroc L, Fairhead C. A new inducible CRISPR-Cas9 system useful for genome editing and study of double-strand break repair in Candida glabrata. Yeast. 2019;36:723–31.
Zhang P, Wang Y, Li C, Ma X, Ma L, Zhu X. Simplified all-in-one CRISPR-Cas9 construction for efficient genome editing in Cryptococcus species. Journal of Fungi. 2021;7:505.
Van Rhijn N, Furukawa T, Zhao C, Mccann BL, Bignell E, Bromley MJ. Development of a marker-free mutagenesis system using CRISPR-Cas9 in the pathogenic mould Aspergillus fumigatus. Fungal Genet Biol. 2020;145:1087–845.
Lax C, Navarro-Mendoza MI, Pérez-Arques C, Navarro E, Nicolás FE, Garre V. Stable and reproducible homologous recombination enables CRISPR-based engineering in the fungus Rhizopus microsporus. Cell Reports Methods. 2021;1:100124.
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Cell biology of infection. In: Molecular Biology of the Cell. New York: Garland Science; 2002.
Amorim-Vaz S, Sanglard D. Novel approaches for fungal transcriptomics from host samples. Front Microbiol. 2016; https://doi.org/10.3389/fmicb.2015.01571.
Chung M, Teigen L, Liu H, Libro S, Shetty A, Kumar N, et al. Targeted enrichment outperforms other enrichment techniques and enables more multi-species RNA-Seq analyses. Sci Rep. 2018;8:1–12.
Watkins TN, Liu H, Chung M, Hazen TH, Dunning Hotopp JC, Filler SG, et al. Comparative transcriptomics of Aspergillus fumigatus strains upon exposure to human airway epithelial cells. Microbial Genomics. 2018;4:e000154.
Lockhart SR, Chowdhary A, Gold JAW. The rapid emergence of antifungal-resistant human-pathogenic fungi. Nat Rev Microbiol. 2023:1–15.
Funding
Open access funding provided by University of the Free State.
Author information
Authors and Affiliations
Contributions
C.C.A. contributed to the study conception and design. C.C.A, A.D., and O.O.K. wrote the main manuscript. C.C.A. prepared figures 1, 2, 3, 4. C.B.N. and R.O. contributed to the editing of the manuscript. All authors reviewed the manuscript and approved the manuscript for publishing.
Corresponding author
Ethics declarations
Conflict of Interest
Conrad Chibunna Achilonu declares that he has no conflict of interest. Adeyinka Davies declares that she has no conflict of interest. Okezie O. Kanu declares that he has no conflict of interest. Colin B. Noel declares that he has no conflict of interest. Rita Oladele declares that she has no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Achilonu, C.C., Davies, A., Kanu, O.O. et al. Recent Advances and Future Perspectives in Mitigating Invasive Antifungal-Resistant Pathogen Aspergillus fumigatus in Africa. Curr Treat Options Infect Dis 16, 14–33 (2024). https://doi.org/10.1007/s40506-023-00269-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40506-023-00269-4