Abstract
Background and Objective
Posaconazole is a pharmacotherapeutic pillar for prophylaxis and treatment of invasive fungal diseases. Dose individualization is of utmost importance as achieving adequate antifungal exposure is associated with improved outcome. This study aimed to select and evaluate a model-informed precision dosing strategy for posaconazole.
Methods
Available population pharmacokinetic models for posaconazole administered as a solid oral tablet were extracted from the literature and evaluated using data from a previously published prospective study combined with data collected during routine clinical practice. External evaluation and selection of the most accurate and precise model was based on graphical goodness-of-fit and predictive performance. Measures for bias and imprecision included mean percentage error (MPE) and normalized relative root mean squared error (NRMSE), respectively. Subsequently, the best-performing model was evaluated for its a posteriori fit-for-purpose and its suitability in a limited sampling strategy.
Results
Seven posaconazole models were evaluated using 764 posaconazole plasma concentrations from 143 patients. Multiple models showed adequate predictive performance illustrated by acceptable goodness-of-fit and MPE and NRMSE below ± 10% and ± 25%, respectively. In the fit-for-purpose analysis, the selected model showed adequate a posteriori predictive performance. Bias and imprecision were lowest in the presence of two prior measurements. Additionally, this model showed to be useful in a limited sampling strategy as it adequately predicted total posaconazole exposure from one (non-)trough concentration.
Conclusion
We validated an MIPD strategy for posaconazole for its fit-for-purpose. Thereby, this study is an important first step towards MIPD-supported posaconazole dosage optimization with the goal to improve antifungal treatment in clinical practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
We evaluated available population pharmacokinetic models for posaconazole from the literature using data from a previously published prospective study combined with data collected during routine clinical practice. |
The model with the best predictive performance was validated for a posteriori fit-for-purpose and its suitability in a limited sampling strategy. |
This study is an important first step towards model-informed precision dosing-supported posaconazole dosage optimization in clinical practice. |
1 Introduction
Posaconazole is a first-line antifungal agent for prophylaxis of various life-threatening invasive fungal diseases [1, 2]. Additionally, posaconazole is deployed for antifungal therapy and was recently found to be non-inferior to voriconazole for the primary treatment of invasive aspergillosis [3]. Posaconazole is highly protein bound, and has a relatively large volume of distribution and an extensive half-life of approximately 35 h [4]. It is primarily eliminated via biliary excretion, and in part by renal excretion and metabolism through glucuronidation [4]. The solid oral tablet is the preferred formulation for oral administration as it demonstrates improved oral bioavailability compared with the suspension formulation [5].
Successful antifungal treatment is highly dependent on effective and safe drug exposure [6, 7]. Therapeutic drug monitoring (TDM) is widely recommended for posaconazole [1]. To achieve optimal individual exposure, model-informed precision dosing (MIPD), an advanced and highly valued tool, can be employed to support dosage optimization.
In MIPD, population pharmacokinetic models are combined with individual patient and disease characteristics, i.e., covariate data, to predict individual exposure through Bayesian forecasting. This can be either performed a priori, in the absence of previous exposure information, for example to select the first dose, or a posteriori, using information from one or more prior concentration measurements, to perform dose adjustments expected to yield optimal exposure [8, 9]. In contrast to classic TDM, MIPD does not require specifically timed concentration measurements and can also be applied using non-trough concentrations [10]. MIPD provides the next-generation approach in antifungal exposure optimization and is an important step forward to ultimately improve antifungal treatment outcomes.
The aim of this study was to perform external validation and fit-for-purpose analyses of the currently available population pharmacokinetic models of posaconazole administered as a solid oral tablet (and intravenous formulation) using a heterogenous adult external evaluation cohort. Thereby, this study aimed to take the first steps towards establishing MIPD for posaconazole and enhance supported decision making for the prevention and treatment of invasive fungal diseases in clinical practice.
2 Methods
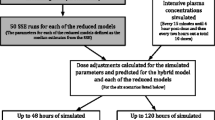
For this study, we used a systematic five-step approach, which is schematically depicted in Fig. 1.
-
Step 1: Identification of suitable population pharmacokinetic models for posaconazole, administered as solid oral tablet, available from the literature.
-
Step 2: Collection of data for the external evaluation cohort from routine clinical care (TDM data) and a previously performed prospective pharmacokinetic study (study data).
-
Step 3: Evaluation of the identified models and selection of the most suitable model using the data from the external evaluation cohort.
-
Step 4: Fit-for-purpose analysis using the final selected model.
-
Step 5: Limited sampling strategy evaluation using the final selected model.
2.1 Model Identification
We performed a structured literature search in PubMed to identify all available parametric population pharmacokinetic models of the solid oral tablet (and intravenous solution) of posaconazole in adult patients until April 2023. The following keywords were used as Medical Subject Heading (MeSH) terms and/or free text for the identification of potential models: ‘posaconazole’ and ‘pharmacokinetic*’. References in the identified articles were screened for any additional relevant publications. We then reviewed articles based on title and/or abstract and included models if they were developed using data from adult individuals receiving the solid oral tablet formulation of posaconazole. Models were excluded if they were non-parametric, based on intravenous data solely, or developed using data from pediatric patients only. Subsequently, we evaluated the remaining full-text articles on suitability before model implementation in the non-linear mixed-effects modeling package NONMEM, version 7.5 (Icon plc, Dublin, Ireland).
2.2 Data Collection
To evaluate the identified models, we collected data for an external evaluation cohort. This external evaluation cohort comprised two sets of data.
For the first dataset, data were retrospectively collected from patients treated with posaconazole and undergoing TDM at the Radboud University Medical Center, Nijmegen, The Netherlands, between 1 December 2014 and 1 December 2021. This dataset is considered a comprehensive representation of a real-world clinical setting. Patients were eligible for inclusion if they were >18 years of age, received posaconazole as solid oral tablet or as intravenous formulation, and had at least one posaconazole plasma concentration measured as part of routine TDM. Both outpatient and clinical patients were included. Data were collected from patients’ medical records and included posaconazole dosing information (e.g., dose, frequency, route of administration), posaconazole TDM information (e.g., plasma concentration, timing of sample collection), patient characteristics (e.g., age, sex, weight, primary underlying disease), and covariate data as defined in the included models. The institutional Medical Ethics Committee waived the necessity to obtain informed consent from these patients.
The second dataset was retrieved from a previously performed prospective observational pharmacokinetic study in critically ill patients receiving posaconazole via intravenous administration. Details on this study, including ethical statement, have been published elsewhere [11, 12]. Contrary to the first dataset, the second dataset not only contained trough concentrations but also information from full pharmacokinetic curves.
All plasma concentrations in this study, both in the TDM and study datasets, were quantified in the same laboratory using a fully validated liquid chromatography-tandem mass spectroscopy (LC-MS/MS) assay. The lower and upper limits of quantification for posaconazole were 0.05 and 10 mg/L, respectively. Plasma concentrations below the limit of quantification were omitted for this analysis. All data in this study were pseudonymized.
For certain covariate data items, namely food status, hematocrit, and albumin and total protein concentrations, information was not widely available in our cohort. To evaluate models including the effect of food status [13,14,15], we assumed that all patients with a hematological malignancy in our cohort were treated under fasting conditions due to their disease state. For all other patients, we assumed posaconazole was administered with food. As albumin concentrations were only missing in non-critically ill patients, we assumed hypoalbuminemia was not present in these individuals. Hematocrit and total protein concentrations were not routinely measured in this cohort. Therefore, we extracted the mean and standard deviation from the values reported in the original publication for imputation in the model for those individuals where these data were missing [16]. We performed a sensitivity analysis by running models with values that were considered at the maximum of realistic to assess the impact of our assumptions on the prediction results. To account for between-occasion variability in pharmacokinetics, occasions in the dataset were defined as described for that specific model. If it was not clearly defined in the article, a new occasion was assumed at every plasma concentration on a different day.
2.3 Model Selection
The identified models including significant covariates were implemented in NONMEM for external evaluation. We obtained model control streams either from the supplementary materials of the original publication or from the information available in the publication. Authors were contacted in the case of uncertainty in coding.
First, we performed the evaluations using the TDM and study dataset separately to assess whether the models would perform differently using these different sets of data. We then performed the model evaluations using the two external evaluation datasets combined. For this part of the analysis, all plasma concentrations were analyzed at the same time. Model evaluation was based on graphical goodness-of-fit and predictive performance [17, 18]. Based on these results, we selected one model to perform a fit-for-purpose analysis.
Graphical goodness-of-fit was assessed by visual inspection of prediction-corrected visual predictive checks (VPCs) [19], individual predicted concentration versus observed concentration plots, and normalized prediction distribution error (NPDE) plots. VPCs aided simulation-based evaluation of appropriateness of the model to describe the concentration-time curve in the external cohort. The NPDE plots visualized the distribution of errors compared with the standard normal distribution N(0,1) [20]. We assessed normality of the NPDE using distribution plots.
The predictive performance of the identified models was assessed by evaluating the models’ ability to predict the observed concentrations. We evaluated this by calculation of the mean percentage error (MPE) as a measure for bias. For this, Eq. 1 was used with N as the total number of observations, predi as the predicted concentration for an individual, and obsi as the observed concentration for an individual.
The root mean squared error (RMSE) and the normalized relative root mean squared error (NRMSE) normalized to the mean of the observed values were calculated as measures for imprecision. For this, Eqs. 2 and 3 were used. We calculated confidence intervals (CIs) for MPE and NRMSE as proposed previously [18, 21].
Additionally, we calculated the fraction of percentage individual prediction errors (PEi) within ± 10% (P10), ± 20% (P20), and ± 30% (P30) to facilitate interpretation. Here, higher values for P10, P20, and P30 can thus be interpreted as favorable. The PEi was calculated using Eq. 4.
2.4 Fit-for-Purpose Analysis
In the a posteriori fit-for-purpose analysis, we assessed the model’s ability to predict a concentration based on the concentration(s) from one or more prior TDM instances using Bayesian forecasting. This analysis was performed with data from the individuals in the external evaluation dataset for whom at least two concentrations were available. For each occasion, we calculated the MPE and NRMSE using Eqs. 1–3. Thereby, this part of the analysis shows the trend in the model’s predictive performance with more prior concentration information becoming available as a posteriori input. Additionally, we evaluated whether predictions of concentrations cause (in)correct (no) dose increases. Four different outcomes were possible:
-
1.
Dose correctly increased: observed and predicted concentrations were both below the target.
-
2.
Dose correctly not increased: observed and predicted concentrations were both above the target.
-
3.
Dose incorrectly increased: observed concentration was above the target, while the predicted concentration was below the target.
-
4.
Dose incorrectly not increased: observed concentration was below the target, while the predicted concentration was above the target.
We used target concentrations of > 0.7 mg/L and > 1.0 mg/L for patients receiving posaconazole as prophylaxis and therapy, respectively [1]. In the absence of a clearly defined relationship between concentration and toxicity, no upper threshold value was used.
2.5 Limited Sampling Strategy Evaluation
In the limited sampling strategy evaluation, we assessed the models’ ability to predict the total drug exposure reflected by the area under the concentration-time curve (AUC) from a single (trough) concentration. This analysis was performed with data from individuals for whom a pharmacokinetic curve was available, i.e., individuals from the prospective study dataset. Here, we used a single pharmacokinetic curve per individual. This approach was chosen because all individuals in this study were critically ill, and it is expected that pharmacokinetics are variable within one individual over time. Using all concentrations from the pharmacokinetic curve, we assessed the AUC by means of post hoc estimation, and assumed this to be the true observed AUC. We then predicted the AUC using only one concentration and compared this with the true observed AUC by calculation of MPE and NRMSE as described earlier. Two approaches for assessing the predicted AUC from a single concentration were performed. First, we used the trough concentration, obtained predose, to calculate the predicted AUC. Hereby, we assessed whether a trough concentration correctly reflected the AUC and could be used as a surrogate for the total posaconazole exposure. Second, we used concentrations obtained at t = 2 (± 1), 4 (± 1), 6 (± 1), or 12 (± 2) h after dose to assess the model’s predictive performance using a single non-trough concentration. Hereby, we evaluated whether the model is suitable for use with non-specifically timed concentration measurements.
3 Results
3.1 Model Identification
A total of seven suitable population pharmacokinetic models describing posaconazole pharmacokinetics after administration of the solid oral tablet (and intravenous formulation) in adult individuals were retrieved from literature and implemented in NONMEM. The PubMed search using the predefined keywords retrieved 346 publications. After evaluation of the title and abstract, 338 publications were excluded as they meet the exclusion criteria. One population pharmacokinetic model was deemed not suitable to be implemented in NONMEM. A summary of the seven identified models as well as the model control streams are presented in the Online Resource. The disposition of posaconazole in the body was described with either one or two compartments. Models differed in the description of the absorption of the solid oral tablet using first-order absorption, sequential zero- and first-order absorption or delayed absorption using transit compartments.
3.2 Data Collection
In the first dataset with retrospectively collected TDM data, 112 patients with 230 evaluable posaconazole plasma trough concentrations were included. Details on this dataset are shown in Table 1. Together with 31 patients and 534 posaconazole plasma concentrations from the previously published prospective pharmacokinetic study [11], the combined external evaluation cohort comprised of 143 patients with 764 posaconazole plasma concentrations with a median (range) trough concentration of 1.65 (0.08–7.02) mg/L.
3.3 Model Selection
The analysis with the retrospective TDM data and the data from the prospective study separately retrieved results, leading to the same conclusions as with the combined dataset. Therefore, only the numeric results and VPCs from the analyses with the separate datasets are shown in the Online Resource, and subsequent analyses were carried forward with the combined dataset.
In Fig. 2, the VPCs of the identified models on the external evaluation data are shown. It can be observed that the data are generally adequately described by most models, but that the Iwasa model fits the data best. This is reflected by the lines for the median, 5th and 95th percentiles of the observed data falling within the shaded areas of the simulated data over the complete concentration-time curve. The Chen (B) model, Petitcollin model, and Van Iersel model seem to generally underpredict the observed concentrations.
Prediction-corrected visual predictive checks of the identified models presented on a linear scale. The observed posaconazole plasma concentrations from the external evaluation datasets are shown as circles; the median, 5th and 95th percentiles of the observed data are shown as solid, dashed, and dashed lines, respectively; and blue shaded areas represent the confidence intervals of the median (dark) and 5th and 95th percentiles (light) of simulated concentrations (n = 500) based on the original dataset
Figure 3 shows the distribution of the NPDE in quantile-quantile (QQ) plots and histograms. For the Iwasa, Jansen, Peña-Lorenzo, and Petitcollin models, the sample quantiles follow the theoretical quantiles, indicated by datapoints in the QQ plots following the theoretical normal distribution line, with slight curve-offs at the extreme values. Additionally, for these models, the distribution of NPDEs are most congruent with that of the overlaid normal distribution in the histograms, indicating accuracy of model predictions. In addition, Fig. 3 shows the individual predicted versus observed concentrations for the identified models. Here, it can be observed that for the Chen (A), Iwasa, Jansen, Peña-Lorenzo, and Petitcollin models, the data are scattered uniformly around the line of unity.
Distribution of the NPDE for the identified models: a quantile-quantile plots of NPDE versus theoretical quantiles of a standard normal distribution N(0,1). b Histograms of NPDE with the density of the standard normal distribution N(0,1) overlaid. c Observed versus predicted posaconazole plasma concentrations for the identified models. NPDE normalized prediction distribution error, N(0,1) normal distribution with a median of 0 and a standard deviation of 1
The results for the predictive performance on the external evaluation cohort are shown in Table 2 and Fig. 4, with MPE, P10, P20 and P30 as measures of bias, and RMSE and NRMSE as measures of imprecision. Based on these results, the Iwasa, Jansen, and Petitcollin models performed best since bias and imprecision were lowest and values were deemed acceptable in light of currently used target concentrations. The results for the sensitivity analysis are presented in the Online Resource. These results support the assumptions we made for those individuals where covariate data were missing.
a MPE and b NRMSE with 95% confidence intervals and percentage of prediction error (c) for the identified models. MPE mean prediction error, NRMSE normalized root mean squared error, P10 percentage of prediction error within ±10%, P20 percentage of prediction error within ± 20%, P30 percentage of prediction error within ± 30%
Based on the above-described results, the Iwasa model was selected as the final model for the fit-for-purpose analysis and limited sampling strategy evaluation.
3.4 Fit-for-Purpose Analysis
We performed the a posteriori fit-for-purpose analysis with data from the individuals from the combined dataset for whom at least two trough concentrations were available. In total, 316 trough concentrations from 85 individuals were available for this part of the analysis.
Figure 5 shows the results of the a posteriori fit-for-purpose analysis describing the model’s ability to predict a concentration based on one or more prior concentration(s). Here, it should be noted that the data became scarcer with an increasing number of occasions. Furthermore, individuals with more observations are expected to show more extreme pharmacokinetics, as more TDM measurements and subsequent dosage adjustments are required to eventually reach adequate exposure. From Fig. 5a, b, it can be observed that bias and imprecision are generally within acceptable values in light of currently used targets for various numbers of previous concentrations available. Fig. 5c indicates that the percentages of correct and incorrect (no) dose increases were quite constant and mostly (approximately 90%) correct. The model predicts a concentration best when two previous concentrations are available, as bias and imprecision were lowest for prediction of the third concentration. Thereby, the model shows potential for the use of dose forecasting in the presence of at least two prior concentrations.
3.5 Limited Sampling Strategy Evaluation
We performed the limited sampling strategy evaluation, using data from 31 individuals from the prospective study with a pharmacokinetic curve available. The pharmacokinetic curves comprised a median (range) of 11 (4–11) samples and were obtained after a median (range) of 2 (2–6) days of posaconazole treatment. In total, 274 plasma concentrations were available for calculation of the true observed AUC. The non-trough concentrations used for calculation of the predicted AUC were obtained after a median (range) of 2.0 (1.5–2.9), 4.0 (3.0–4.8), 6.0 (5.5–6.8), and 12.0 (10.2–13.8) h after dose administration and were available for 30, 26, 24, and 28 individuals, respectively.
Figure 6 shows the observed versus predicted total posaconazole exposure from the individuals in the limited sampling strategy evaluation. Generally, most datapoints were close to the line of unity. Numerical results for this part of the analysis are available via Table 3. When predicting the AUC using a single trough concentration after intravenous administration, the calculated MPE and NRMSE (95% CI) were 10.3% (2.7–17.8%) and 18.2% (13.3–22.9%), respectively. When predicting the AUC using a single non-trough concentration after intravenous administration, the calculated MPE and RMSE were all within ±6.2% and ±35.3%, respectively.
Observed versus predicted posaconazole exposure for the limited sampling strategy evaluation using the selected final model with a trough and b non-trough concentrations. The results for the analysis with non-trough concentrations collected at 2 (± 1), 4 (± 1), 6 (± 1), and 12 (± 2) h after dose are shown. AUC area under the concentration-time curve
4 Discussion
We validated a clinical MIPD strategy for posaconazole administered as a solid oral tablet or intravenous formulation in a heterogenous adult patient population. With our analyses, we have taken an important step forward to translate population pharmacokinetics of posaconazole from bench to bedside. Thereby, this study supports posaconazole dose decision making with the ultimate goal of improving treatment outcomes for patients with invasive fungal diseases.
We demonstrated that multiple models performed accurately and precisely in describing observed posaconazole pharmacokinetics. The model by Iwasa et al. [14] showed the best predictive performance and was selected for subsequent fit-for-purpose analyses. A potential explanation for its predictive performance might be that this model was developed based on a heterogenous population, including healthy volunteers and patients with various underlying diseases. This is congruent with our external evaluation cohort. Additionally, from all the models we evaluated, the Iwasa model was developed based on the largest number of individuals with a wide age and weight range and concentrations over a wide time-after-dose range [14]. Such models are expected to be more generalizable to external populations than models developed with data from a small set of patients with trough concentrations only [9]. This is consistent with findings from external evaluation studies for other drugs [10].
When multiple models are available for one drug, the selection of a model for MIPD ideally matches the target population [9]. As posaconazole is a drug deployed in a heterogeneous patient population, including but not limited to critically ill patients, hematology patients, and patients with chronic lung diseases, the selected model would ideally show good predictive performance in this various population. Since these different patient populations display variations in posaconazole pharmacokinetics, another approach could be to select a model for each target population. Our analyses in two subsets of data did not lead to different conclusions for model selection. This indicates appropriateness of the use of the selected model for MIPD in a clinical setting. Future studies could aid in confirming the predictive performance of this model in a prospective validation study.
Our external evaluation cohort consisted partially of retrospectively collected data that potentially introduced bias [22]. For multiple models, the predictive performance for posaconazole plasma concentrations at time after dose exceeding 24 h was inadequate. This might be caused by incorrect registration of last dose intake before sample collection. Combining our retrospective dataset, a comprehensive representation of a real-world clinical setting, with a prospectively collected dataset is considered to improve the overall quality of the data we used [11]. Another challenge regarding data used in this study was that not all covariate data were available. To overcome this, we calculated the mean and standard deviation of the covariates reported in the original publication and used this to impute the missing values in our dataset. We performed a sensitivity analysis that showed absence of clinically relevant impact on the predictive performance of those models where it was needed to make these assumptions, i.e., the models by Chen (B), Jansen, and Peña-Lorenzo [23,24,25]. Moreover, we are aware of the significant impact that certain coadministered drugs have on posaconazole pharmacokinetics through drug–drug interactions [26]. Data on drug–drug interactions were however not included in our analysis since none of the identified models included the presence of a coadministered drug as a significant covariate in their final model.
Recently, Huang et al. performed an evaluation study of the predictive performance of population pharmacokinetic models for posaconazole [27]. The authors concluded that none of the evaluated models showed adequate predictive performance for their cohort. They attributed this to the difference between Caucasian and Chinese patients, of whom data were used for model development and external evaluation, respectively. Regrettably, the Iwasa and Chen (B) models, based in part on data from Japanese and Chinese patients, were not evaluated in the previous study, nor were the Chen (A), Jansen and Van Iersel models. Huang et al. included models developed on intravenous data only and used oral data as input for their external evaluation [27]. We are convinced that oral data should not be used in an intravenous model, as this completely ignores the absorption process of the orally administered drug and assumes 100% oral bioavailability. Their conclusion of poor predictive performance was probably impacted by their study design. Contrary to the study by Huang et al. [27], we focused on models based on solid oral tablet formulation data with or without intravenous data combined. Additionally, Huang et al. evaluated pediatric models using adult data [27]. We excluded models developed with data from pediatric populations solely as we deemed it inappropriate to perform model evaluations in the absence of extensive pediatric data. Taking the above into account, we consider our study of added value. On top of this, we performed fit-for-purpose analyses and limited sampling strategy evaluations showing the effect in dose decision making and AUC prediction from a single (trough) concentration. Thereby, we showed the clear advantages of the selected MIPD strategy for its use in clinical practice.
In the fit-for-purpose analysis, we validated the Iwasa model for its use for MIPD of posaconazole in clinical practice. The model predictions resulted in correct dose increase decisions in almost all cases. As no clear posaconazole exposure-toxicity relationship has been identified to date, we validated dose increase decisions only. Although an upper threshold concentration of 3.75 mg/L has been proposed based on the 90th percentile of observed average concentrations after administration of the oral suspension formulation [4], this approach was chosen because higher concentrations are also shown to be tolerated [28]. The model accurately predicted posaconazole trough concentration with information from one or more prior concentration measurements. It should however be noted that it seems necessary to inform the model with at least two concentrations to precisely predict the next concentration. This is reflected by greater imprecision, with only one prior measurement being available. These findings suggest that dose decisions based on this model should be verified with follow-up posaconazole concentration measurements to reduce imprecision. This underlines the attributed value of repeated assessment of posaconazole exposure. In our analyses, we weighted each observation equally. It could be hypothesized that for critically ill patients, this may not be the ideal approach given the expected variability in pharmacokinetics and thus in concentrations. This could be considered a limitation of our analyses, but we deem it beyond the scope of our study to analyse the weighting of observations.
It is generally assumed that posaconazole plasma trough concentrations are a good surrogate for the overall posaconazole exposure reflected by the AUC [29]. For clinical practice, the assessment of a trough concentration poses a minimally invasive method associated with less costs compared with assessment of the AUC. We performed an analysis to evaluate the suitability of the selected model to be used in such a limited sampling strategy. Our findings show accurate and precise model-derived AUC predictions, confirming the assumption that a posaconazole trough concentration is a good surrogate for a posaconazole AUC. In addition, we validated this model’s predictive performance using non-trough concentrations showing acceptable bias and imprecision. Thereby, we confirmed that the selected model for MIPD of posaconazole does not require specifically timed concentration measurements showing a clear advantage compared with traditional TDM. This analysis was performed using only intravenous data as all patients with full pharmacokinetic curves received posaconazole intravenously and should be interpreted in that light. Future studies using data after oral administration with rich sampling should confirm whether this also holds for exposure predictions after oral administration.
5 Conclusion
With this study, we took an important step to bring MIPD for posaconazole into clinical practice. Thereby, this study aids in improving individual exposure of posaconazole, and subsequently contributes to personalized antifungal treatment for patients at risk for and with invasive fungal diseases.
References
Ullmann AJ, Aguado JM, Arikan-Akdagli S, Denning DW, Groll AH, Lagrou K, et al. Diagnosis and management of Aspergillus diseases: executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clin Microbiol Infect. 2018;24(Suppl 1):e1–38.
Maertens JA, Girmenia C, Bruggemann RJ, Duarte RF, Kibbler CC, Ljungman P, et al. European guidelines for primary antifungal prophylaxis in adult haematology patients: summary of the updated recommendations from the European Conference on Infections in Leukaemia. J Antimicrob Chemother. 2018;73(12):3221-30.
Maertens JA, Rahav G, Lee DG, Ponce-de-Leon A, Ramirez Sanchez IC, Klimko N, et al. Posaconazole versus voriconazole for primary treatment of invasive aspergillosis: a phase 3, randomised, controlled, non-inferiority trial. Lancet. 2021;397(10273):499–509.
European Medicines Agency. 2014. Noxafil. Assessment Report. https://www.ema.europa.eu/en/documents/variation-report/noxafil-h-c-610-x-0028-epar-scientific-discussion-extension_en.pdf. Accessed 1 Aug 2023.
Krishna G, Ma L, Martinho M, Preston RA, O’Mara E. A new solid oral tablet formulation of posaconazole: a randomized clinical trial to investigate rising single- and multiple-dose pharmacokinetics and safety in healthy volunteers. J Antimicrob Chemother. 2012;67(11):2725–30.
Jang SH, Colangelo PM, Gobburu JV. Exposure-response of posaconazole used for prophylaxis against invasive fungal infections: evaluating the need to adjust doses based on drug concentrations in plasma. Clin Pharmacol Ther. 2010;88(1):115–9.
Walsh TJ, Raad I, Patterson TF, Chandrasekar P, Donowitz GR, Graybill R, et al. Treatment of invasive aspergillosis with posaconazole in patients who are refractory to or intolerant of conventional therapy: an externally controlled trial. Clin Infect Dis. 2007;44(1):2–12.
Jelliffe RW, Schumitzky A, Van Guilder M, Liu M, Hu L, Maire P, et al. Individualizing drug dosage regimens: roles of population pharmacokinetic and dynamic models, Bayesian fitting, and adaptive control. Ther Drug Monit. 1993;15(5):380–93.
Keizer RJ, Ter Heine R, Frymoyer A, Lesko LJ, Mangat R, Goswami S. Model-informed precision dosing at the bedside: scientific challenges and opportunities. CPT Pharmacometr Syst Pharmacol. 2018;7(12):785–7.
Wicha SG, Martson AG, Nielsen EI, Koch BCP, Friberg LE, Alffenaar JW, et al. From therapeutic drug monitoring to model-informed precision dosing for antibiotics. Clin Pharmacol Ther. 2021;109(4):928–41.
Van Daele R, Wauters J, Dreesen E, Boelens J, Nulens E, Lormans P, et al. Exposure to intravenous posaconazole in critically ill patients with influenza: a pharmacokinetic analysis of the POSA-FLU study. Mycoses. 2022;65(6):656–60.
Vanderbeke L, Janssen NAF, Bergmans D, Bourgeois M, Buil JB, Debaveye Y, et al. Posaconazole for prevention of invasive pulmonary aspergillosis in critically ill influenza patients (POSA-FLU): a randomised, open-label, proof-of-concept trial. Intensive Care Med. 2021;47(6):674–86.
Chen L, Krekels EHJ, Heijnen AR, Knibbe CAJ, Bruggemann RJ. An integrated population pharmacokinetic analysis for posaconazole oral suspension, delayed-release tablet, and intravenous infusion in healthy volunteers. Drugs. 2023;83(1):75–86.
Iwasa T, de Almeida C, Fauchet F, Winchell GA, de Greef R, Hasegawa C, et al. Model-informed dose justifications of posaconazole in Japanese patients for prophylaxis and treatment against fungal infection. J Clin Pharmacol. 2023;63(4):421–34.
van Iersel M, Rossenu S, de Greef R, Waskin H. A population pharmacokinetic model for a solid oral tablet formulation of posaconazole. Antimicrob Agents Chemother. 2018;62(7).
Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135.
Ette EI, Williams PJ, Kim YH, Lane JR, Liu MJ, Capparelli EV. Model appropriateness and population pharmacokinetic modeling. J Clin Pharmacol. 2003;43(6):610–23.
Sheiner LB, Beal SL. Some suggestions for measuring predictive performance. J Pharmacokinet Biopharm. 1981;9(4):503–12.
Bergstrand M, Hooker AC, Wallin JE, Karlsson MO. Prediction-corrected visual predictive checks for diagnosing nonlinear mixed-effects models. AAPS J. 2011;13(2):143–51.
Brendel K, Comets E, Laffont C, Mentre F. Evaluation of different tests based on observations for external model evaluation of population analyses. J Pharmacokinet Pharmacodyn. 2010;37(1):49–65.
Faber NKM. Estimating the uncertainty in estimates of root mean square error of prediction: application to determining the size of an adequate test set in multivariate calibration. Chemometr Intell Lab. 1999;49(1):79–89.
Mu S, Ludden TM. Estimation of population pharmacokinetic parameters in the presence of non-compliance. J Pharmacokinet Pharmacodyn. 2003;30(1):53–81.
Chen L, Krekels EHJ, Dong Y, Chen L, Maertens JA, Blijlevens NMA, et al. Meta-PK analysis of posaconazole upon dosing of oral suspension, delayed-release tablet, and intravenous infusion in patients versus healthy volunteers: impact of clinical characteristics and race. Int J Antimicrob Agents. 2023;62(6): 106995.
Jansen AME, Muilwijk EW, van der Velden W, Maertens JA, Aerts R, Colbers A, et al. Posaconazole bioavailability of the solid oral tablet is reduced during severe intestinal mucositis. Clin Microbiol Infect. 2022;28(7):1003–9.
Pena-Lorenzo D, Rebollo N, Sanchez-Hernandez JG, Zarzuelo-Castaneda A, Vazquez-Lopez L, Otero MJ, et al. Population pharmacokinetics of a posaconazole tablet formulation in transplant adult allogeneic stem cell recipients. Eur J Pharm Sci. 2022;168: 106049.
Bruggemann RJ, Alffenaar JW, Blijlevens NM, Billaud EM, Kosterink JG, Verweij PE, et al. Clinical relevance of the pharmacokinetic interactions of azole antifungal drugs with other coadministered agents. Clin Infect Dis. 2009;48(10):1441–58.
Huang S, Ding Q, Yang N, Sun Z, Cheng Q, Liu W, et al. External evaluation of published population pharmacokinetic models of posaconazole. Front Pharmacol. 2022;13:1005348.
Schauwvlieghe A, Buil JB, Verweij PE, Hoek RAS, Cornelissen JJ, Blijlevens NMA, et al. High-dose posaconazole for azole-resistant aspergillosis and other difficult-to-treat mould infections. Mycoses. 2020;63(2):122–30.
Bruggemann RJ, van Luin M, Colbers EP, van den Dungen MW, Pharo C, Schouwenberg BJ, et al. Effect of posaconazole on the pharmacokinetics of fosamprenavir and vice versa in healthy volunteers. J Antimicrob Chemother. 2010;65(10):2188–94.
Acknowledgements
The authors thank Anne van Schip for her contributions to this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
Roger J.M. Brüggemann has served as consultant to Astellas Pharma, Inc., F2G, Amplyx, Gilead Sciences, Merck Sharp & Dohme Corp., Mundipharma, and Pfizer, Inc., and has received unrestricted research grants from Astellas Pharma, Inc., Gilead Sciences, Merck Sharp & Dohme Corp., and Pfizer, Inc. All contracts and payments were through his institution. Ron J. Keizer is an employee and stockholder of InsightRX, a company developing precision dosing software. Anouk M.E. Jansen, Kim Snijdelaar, Isabel Spriet, Erwin Dreesen, and Rob ter Heine have no conflicts of interest to declare in relation to the current work.
Funding
No funding or sponsorship was received for this study or publication of this article.
Data availability statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request and following legal approval.
Ethical approval
The Medical Ethics Committee of the Radboud University Medical Center, Nijmegen, The Netherlands, waived the necessity to obtain informed consent from the patients for whom data were retrospectively collected. The ethical approval statement for the previously performed prospective study has been published elsewhere [11, 12].
Authors’ contributions
AJ, RB, and RtH conceptualized and designed the study; AJ and KS performed data collection; AJ performed the analyses and interpreted the results with RtH and RB; and AJ wrote the first draft of the manuscript. All authors critically reviewed the manuscript and approved the final version.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Model codes are available through the Supplementary Materials.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Jansen, A.M.E., Snijdelaar, K., Keizer, R.J. et al. Personalized Antifungal Therapy Through Model-Informed Precision Dosing of Posaconazole. Clin Pharmacokinet 63, 645–656 (2024). https://doi.org/10.1007/s40262-024-01361-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-024-01361-8