Abstract
HCV is a hepatotropic RNA virus recognized for its frequent virulence and fatality worldwide. Despite many vaccine development programs underway, researchers are on a quest for natural bioactive compounds due to their multivalent efficiencies against viral infections, considering which the current research aimed to figure out the target-specificity and therapeutic potentiality of α, β, and δ subunits of amyrin, as novel bioactive components against the HCV influx mechanism. Initially, the novelty of amyrin subunits was conducted from 203 pharmacophores, comparing their in-silico pharmacokinetic and pharmacodynamic profiles. Besides, the best active site of CD81 was determined following the quantum tunneling algorithm. The molecular dynamic simulation was conducted (100 ns) following the molecular docking steps to reveal the parameters- RMSD (Å); Cα; RMSF (Å); MolSA (Å2); Rg (nm); PSA (Å); SASA (Å2), and the MM-GBSA dG binding scores. Besides, molecular strings of CD81, along with the co-expressed genes, were classified, as responsible for encoding CD81-mediated protein clusters during HCV infection, resulting in the potentiality of amyrins as targeted prophylactics in HCV infection. Finally, in vivo profiling of the oxidative stress marker, liver-specific enzymes, and antioxidant markers was conducted in the DMN-induced mice model, where β-amyrin scored the most significant values in all aspects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis C Virus (HCV) is a small, single-stranded, positive-sense RNA virus identified in 1989 and belongs to the Hepacivirus genus of the Flaviviridae family (Lavanchy 2011). According to World Health Organization (WHO), around 71 million people are infected, and about 400,000 individuals die due to complications caused by long-term HCV infections (Morozov and Lagaye 2018). HCV infects the hepatic cells, leading to inflammation (Zaltron et al. 2012). Among the HCV patients, 25% experience acute infection (Grebely et al. 2012), whereas 75% suffer from progressive liver failure, cirrhosis, and hepatocellular carcinoma (Chen and Morgan 2006). Infection by HCV may also lead to extrahepatic diseases like diabetes mellitus, Non-Hodgkin's lymphoma, membranoproliferative glomerulonephritis, and lichen planus. It can significantly reduce the quality of life of the infected individuals (Millman et al. 2017). HCV comes in various genotypes, including- 1a, 1b, 3a, 3b, 4, and 6, with 3a being the most virulent form (Kalinina 2015). HCV RNA (9.5 kb) encodes a single open reading frame (ORF) (Wu et al. 2015). This ORF translates into a polyprotein consisting of 3000 amino acids (Pfaender et al. 2014). This polyprotein processes into envelopes (E1 and E2) and seven nonstructural proteins (Bruening et al. 2018). The structural HCV proteins are Core, E1, E2, and p7, which divide structural proteins from nonstructural proteins (Morozov and Lagaye 2018). The interaction of HCV with hepatocytes leads to viral entry depending on host lipoprotein components and viral envelope glycoproteins. Here, host factors are present at the surface of the hepatocytes. The virus enters hepatocytes via interactions between the viral envelope proteins- E1 and E2 and four known host receptors (CD81, Claudin-1, SBR-I, and occluding) (Ploss et al. 2009). After entry, host ribosomes bind to the internal ribosomal entry site (IRES) of the HCV genome and translate viral polyproteins on the rough endoplasmic reticulum (ER) (Moradpour et al. 2007). Host and viral proteases produce the polyprotein into two components. One is the structural (core and envelope proteins- E1 and E2) and nonstructural proteins (p7, NS2–3, NS3, NS4A, NS4B, NS5A, and NS5B) (Lindenbach and Rice 2005). Glycoproteins E1 (35 kDa) and E2 (70 kDa) are spliced from the precursor in the endoplasmic reticulum by the host's cellular signal peptidase. The glycoproteins E1 and E2 have ectodomains that are highly glycosylated. The E1 has about 4 to 5, and the E2 has 11 N-glycosylation regions (Morozov and Lagaye 2018). E1 and E2 ectodomains contain cysteine residues in their structure at positions 8 and 18. It forms intra-molecular and inter-molecular disulfide bonds (Chigbu et al. 2019). That adheres the envelope proteins (E1, E2) to the host surface protein. Hypervariable region 1 HRV1) and hypervariable region 2 (HRV2) are two highly transmutable areas found in E2 (Prentoe et al. 2016). These areas aid HCV in evading the immune system and promoting hepatic cell infection. E2 contains multiple invariant areas that aid in antibody neutralization and play a vital role in the HCV's lifespan (Lavie et al. 2014).
The CD81 receptor protein is the main corridor of E2 receptor binding and entry (Sharif et al. 2021), belongs to the tetraspanin (transmembrane 4) family of proteins, traverses the membrane four times, and has two extracellular (EC) loops of 28 and 80 aa, designated as- EC1 and EC2, respectively (Levy et al. 1998). It weighs 26 kDa and contains- a large extracellular loop (LEL), a small extracellular loop (SEL), an intracellular N- and c-terminal domain, and four membrane-spanning domains. E2 binds to the LEL region of CD81 effectively (Burlone and Budkowska 2009). The engagement of CD81 activates various biologic responses, including cell adhesion, morphology, proliferation, activation, and differentiation of T, B, and other cell types (Levy et al. 1998). HCV interaction with CD81 I induces multiple downstream signaling pathways, including Rho GTPase family members, Cdc42, and mitogen-activated protein kinase pathways (Farquhar et al. 2011). EFGR is a coenzyme that aids in developing the CD81-CLDN1 complex and upregulates several downstream molecules in preparation for HCV genetic material insertion (Li et al. 2021). Characterization of CD81 is through four conserved cysteine residues, including a ubiquitous CCG motif, and two additional cysteines in the LEL, that form critical disulfide bonds in LEL. Other regions of CD81, especially- the C-terminal region, transmembrane residues, and post-translational processing, also play crucial roles in the HCV influx mechanism through the alternative pathways (Zona et al. 2014). The residues of the transmembrane domains and cysteine-mediated palmitoylation exhibit moderate inhibitory effects on HCV entry (Fénéan et al. 2014). It indicates that the CD81 LEL chain is one of the potent critical determinants of viral entry (Asselah et al. 2009).
Although several vaccine candidates have gone through a developmental phase, the genetic variation and high level of adaptability of the virus make it challenging to find an effective treatment for HCV infection (Ansaldi et al. 2014). In pharmaceutical sciences, plant-derived has garnered much attention as potential prophylactic-therapeutic agents against infectious diseases and immunological disorders (Jardim et al. 2018). The World Health Organization (WHO) announced a phytochemicals control strategy for HCV infection (Kumar et al. 2020). Plant-derived phytocompounds have a wide range of physiologically active properties, such as antiviral, anti-inflammatory, antifungal, and antibacterial actions, with the least possible side effects reported. Several phytoactive substances naturally resist HCV (Patel et al. 2021). Amyrin subunits have the properties and potentiality to suppress protease inhibitors-resistant HCV infection by preoccupying the CD81 receptors (Oh et al. 2021). The cannabinoid receptors- CB1, CB2; the production of cytokines; the expression of NF-KB, CREB; and cyclooxygenase-2 have long-lasting antinociceptive and anti-inflammatory properties in both the 'α and β-Amyrins.' Amyrin subunits are hyperactive in silico pharmacokinetics and pharmacodynamics properties. As a result, they are promising candidates for HCV treatment (Zhang et al. 2022).
Considering the aforementioned factors, the current study aimed to analyze the profiles of the CD81 receptor in terms of quantum tunnels; molecular-string networks; and superactive positioning so that the target specificity of different amyrin subunits can be determined as novel bioactive compounds following the molecular docking and dynamic simulation approaches. Besides, the in-vivo study was conducted comprehensively to validate the findings of the pharmacophores used before undertaking any clinical applications.
Materials and methods
Library construction of the phytochemical compounds
A library of 203 phytochemical compounds was constructed using the interface of PubChem (https://pubchem.ncbi.nlm.nih.gov/) and, ChEMBL (https://www.ebi.ac.uk/chembl/) database. The contemporary established literature reviews comprise- comprehensive clinical and nonclinical studies. Also, mentions the standard accepted drugs globally and functional bioactive agents (i.e., flavonoids, isoflavonoids, carotenoids, isoprenoids, phytosterols, limonoids, etc.).
Pharmacokinetic analysis using ADMET and QSAR profiling
The compounds' pharmacokinetic properties determine the compounds' disposition inside the human body (Duran et al. 2020). To understand the absorption, distribution, metabolism, and excretion, server tools such as Molinspiration (https://www.molinspiration.com/cgi-bin/properties) and 'Swiss ADME' (http://www.swissadme.ch/index.php) were used as preliminary (Ferdausi et al. 2022). Afterward, 'admetSAR 2.0' (http://lmmd.ecust.edu.cn/admetsar2/) was used for secondary verification purposes. Finally, the toxicity profiles of the compounds were conducted through 'pkCSM' (http://biosig.unimelb. edu.au/pkcsm/prediction) (Dey et al. 2021). In the case of ADMET and QSAR analysis, parameters like- molecular weight, number of the hydrogen bond acceptor and donor, predicted octanol/water partition coefficient, number of rotatable bonds, intestinal absorption, total clearance, LD50, blood–brain barrier, hepatotoxicity, AEMS toxicity, maximum human tolerant doses, drug-likeness and number of Lipinski's rule violations, were considered (Nipun et al. 2021). The QSAR (quantitative structure–activity relationship) analysis of the desired ligands was analyzed using the PASS server (http://www.way2drug.com/passonline/) to finally validate the anti-microbial, antiviral, and anti-infective characteristics (Arefin et al. 2021).
Selection of the receptor macromolecule
The crystal structure of the human 'Cluster of Differentiation 81 (CD81)' protein (PDB ID: 1G8Q) as the prime receptor for hepatitis C virus (HCV) was selected and downloaded from the protein data bank (https://www.rcsb.org/) in PDB format (Arefin et al. 2021), where the X-ray diffraction resolution was 1.60 Å. The FASTA format of the CD81 receptor macromolecule was run in the 'SEQATOMs' web interface (https:// www.bioinformatics.nl/tools/seqatoms/) to detect the missing residues of the crystal structure of the protein (Brandt et al. 2008). More importantly, the absent residues of the protein's resolved structure's middle, C, and N terminus regions were assessed by the BLAST function of SEQATOMs (Brandt et al. 2008).
Optimization of components
Optimization of receptor protein
The crystal structure of the receptor macromolecule- CD81 (PDB ID: 1G8Q) was optimized using 'UCSF Chimera version 1.14' (https://www.cgl.ucsf.edu/chimera/download.html) (Arefin et al. 2021). Here, non-interactive residues, ions, water molecules, unwanted ligands, and side chains except the 'A chain' of CD81 were removed. Besides, the missing hydrogen atoms are added (Fig. 1A). 'Gasteiger method' for energy minimization of the protein, and the output file was saved as a 'pdb' format. Finally, the quantitative measurement of the minimized energy was accomplished through the 'YASARA' (http://www.yasara.org/minimizationserver.htm) (Arefin et al. 2021).
Optimization of ligands
The structures of the test ligands of interest mean- α, β, and δ-Amyrin (PubChem CID: 73,170, 73,145, 12,358,447, respectively) were downloaded as SDF files from PubChem, following their ADMET and QSAR profiles, where Benzyl salicylate (PubChem CID: 8363) was used as the control (Fig. 1B). The accumulative charge of the ligands up to zero is removed, the energy minimization process regulates as an essential part of optimization, following the Gasteiger method of 'UCSF Chimera version 1.14' (Yang et al. 2013). After energy minimization, each test ligand was kept as a 'mol2 file' (Fig. 1C–E).
Active site prediction of the CD81 receptor protein
Here, point-specific molecular docking is performed. The active sites of the receptor were initially identified using the 'CASTp Server' (http://sts.bioe.uic.edu/castp/) (Tian et al. 2018), where only a single region of 95.271 Å surface area was spotted. Secondary identification of the active sites was accomplished using 'Maestro-Schrödinger' (https://www.schrodinger.com/products/desmond), where four different active sites have resulted. The high-throughput prediction of the exposed and hidden active sites of CD81 using the 'COACH-D' (https://yanglab.nankai.edu.cn/COACH-D/) algorithm was conducted finally (Yang et al. 2013) to verify the authenticity of those sites identified by 'Maestro-Desmond.' The 'COACH-D' algorithm revealed- the four active sites (Fig. 2A–D) along with the nearby amino acid residues, their positional strength, and binding energy (Kcal/mol) simultaneously and also in comparison to each other (Fig. 2E).
Quantum tunneling on the best active site of CD81
To assess the morphological features of the best active site (Fig. 2D) of the CD81 receptor, quantum tunneling parameters were analyzed considering- the protein tunnel length (Å), curvature (radius), and bottleneck (radius). In that case, three different protein tunnels were identified (Fig. 3A–C), each containing seven sub-tunnel clusters at various configurations (Fig. 3D–F). In all the parameters, the third tunnel (Fig. 3C) is more viable than all the others, along with the sub-tunnel clusters it possesses (Fig. 3F). Thus, the third tunnel region of the active site of CD81 predicts the supramolecular docking point for any ligands. In all aspects of quantum tunneling of CD81, 'CAVER 3' (http://www.caver.cz) (35; Kingsley and Lill 2014), and 'Site Map 2.6' (Schrödinger, LLC) were used (John et al. 2015).
Demonstration of the three major protein tunnels of the CD81 receptor (A–C) qualitatively resulted from the quantum mechanics of tunnel formation using the CAVER 3.0 tool. Besides, seven sub-tunnels of each of the three major tunnels were analyzed quantitatively depending on their length (D), curvature radius (E), and bottleneck radius (F)
Point-specific molecular docking
Individual molecular docking of each of the optimized ligands is done, keeping the best active site of the CD81 receptor into account through Maestro (Schrödinger, LLC) (Lyne et al. 2006). At the molecular docking operation, the macromolecule and the ligands convert into 'pdbqt file' format. The RMSD values assessed (Å) and the binding affinities of each ligand–protein complex were conserved in 'CSV' format for further studies.
Post molecular docking analysis
The qualitative analysis and initial visualization of the ligand–protein complexes were undertaken by 'PyMOL version 2.5 (https://pymol.org/2/) and 'Discovery Studio Visualizer version 3.0' (https://discover.3ds.com/discovery-studio-visualizer-download). The ligand–protein complex files were saved in 'pdb' format each time. As the secondary study, a quantitative assessment of the number of hydrophobic interactions (non-covalent) and the number of hydrogen bond formation between each ligand-receptor complex was studied through 'LigPlot + version 2.2' (Dey et al.2021; Nipun et al. 2021; Arefin et al. 2021), before subjecting into the molecular dynamic simulation (MDS).
Molecular dynamic simulation (100 ns)
At the very beginning, the physical alterations and frequent interactions of the ligand-free CD81 receptor with the neighboring water molecules and ions were observed for 10 ns, operating the 'CABS-flex 2.0' web-based simulator (http://biocomp.chem.uw.edu.pl/CABSflex2/) (Kuriata et al. 2018). Afterward, the protein–ligand complexes were simulated up to 3.1 ns in the 'LARMD' simulation system (http://chemyang.ccnu.edu.cn/ccb/server/LARMD/index.php). It helps in understanding the Debye–Waller factor for thermostability (B-factor), residual cross-correlations (RCC), and a clustering dendrogram from the principal component analysis (Yang et al. 2020). Then a comprehensive molecular dynamic simulation of the ligand–protein complexes was conducted by 'Desmond Simulation Package' (Schrödinger, LLC, NY, USA) for 100 ns to analyze critical parameters- RMSD (Å), Cα, RMSF (Å), MolSA (Å2), Rg (nm), PSA (Å), SASA (Å2), and the MM-GBSA dG binding score (Ivanova et al. 2018). The grid box dimension was fixed at 'X: Y: Z', and Na + were added as the nullifying ion required to get the expected results. Here, assessment of the SASA and MolSA values, the probe radius was adjusted to 1.4 Å. All the resulting data were converted and conserved into 'CSV' format.
CD81-mediated genetic string networking to determine the protein clustering
The CD81 gene-mediated string networks assess the coexpressed and overexpressed genes responsible for controlling the CD81 protein synthesis in living systems. Firstly, the CD81 receptor protein string was extracted from the STRING database (https://string-db.org/). The individual nodes and edges of the protein were identified and characterized using Cytoscape 3.8.2 (https://cytoscape.org/) (Li et al. 2017), which operates through the Java Runtime Environment (https://www.oracle.com/java/technologies/downloads/). Secondly, the genes responsible for encoding the CD81 receptor and the others significantly interconnected with the CD81 protein were screened using GeneMANIA (https://string-db.org/) (Warde et al. 2010). The protein clustering was accomplished considering- nodes of string formation; neighborhood on the chromosome; gene-fusion; phylogenetic co-occurrence; homology; coexpression; experimentally determined interactions; database annotation; automated text mining, and the combined scores. The clustering of the interactive CD81 nodes was constructed with the tools- Morpheus (https://software.broadinstitute.org/morpheus) (Molinari et al. 2021) and Heatmapper web interface (http://www2.heatmapper.ca/expression/) (Babicki et al. 2016). Finally, the CD81 connected-gene clusters responsible for entering the viral particles (E2 protein of HCV) inside the host were identified by GeneMANIA (https://string-db.org/) (Warde et al. 2010).
DMN-induced modeling of hepatic fibrosis in mice
In this study, Swiss albino mice were modeled for studying dimethylnitrosamine (DMN)-induced hepatic fibrosis study (Thirupathi et al. 2017), grouping them into control group (CG), negative control group (NCG), treatment group-1 (TG-1) and treatment group-2 (TG-2) (Al Azad et al. 2020a). In the NCG, DMN was administrated, whereas TG-2 was given both the DMN and β-amyrin. In contrast, CG and TG-1 were totally free from DMN. Oxidative-stress and antioxidant markers were also studied comprehensively (Table 7) such as Vit. C (mmol/ml), CAT (µmol/ml), GSH (mg/ml), GPX (mg/ml), SOD (U/ml), and MDA (nmol/ml). In addition, liver-specific serological parameters mean alkaline phosphatase, ALP (IU/L), lactate dehydrogenase, LDH (IU/L), aspartate aminotransferase, AST (IU/L), alanine transaminase, ALT (IU/L) were analyzed (Table 8) (Thirupathi et al. 2017).
Statistical analysis and graphical representation
The resulting data from the molecular dynamic simulation (100 ns) and the aforementioned in vivo steps were statistically analyzed using 'R programming' (version R-4.0.2 for Linux) (Akter et al. 2020; Islam et al. 2021; Paul et al. 2022; Morshed et al. 2022), and 'GraphPad Prism version 8.0.1' software package (for Mac OS) (Azad et al. 2020a, b; Rashaduzzaman et al. 2019; Al Azad et al. 2022).
Results
Detection of protein active site
In the CD81 receptor, four active sites have been identified (Fig. 2), where the fourth one is predicted to be the most active site (Fig. 2D) for molecular docking. According to the number of amino acids residues involved in that location and the expected restored energy level (Fig. 2E). In contrast, the rest of the active sites (Fig. 2A–C) were found to be inferior to that of the fourth one in all aspects (Fig. 2E). Finally, the quantum tunneling algorithm was applied only on the fourth active (Fig. 2D) site in characterizing the location for super molecular docking.
Quantum mechanics for protein tunneling
A Quantum tunneling algorithm applies to the best active position (Fig. 2D) of the CD81 receptor, from which three different protein tunnels were identified (Fig. 3A-C). Interestingly, each of these tunnels contains a cluster of seven sub-tunnels (Fig. 3D–F). Each of the significant tunnels, along with the sub-tunnel clusters, was distinctive and unique to each other, considering the parameters such as tunnel length (Å), curvature (radius), and bottleneck radius (radius). It's quite transparent that, the number of amino acids near the bottleneck point of each of the significant tunnels are different from each other, meaning- Thr166, Thr167, Asn180, Leu185, Asn184, Cys157, and 163Thr were observed in the first considerable tunnel (Fig. 3A). In the same way, Cys156, Asp189, Cys190, Ser160, Gly158, Thr166, and Asn184 are observed in the second central tunnel (Fig. 3B). In contrast to these two tunnels, the length and curvature of the third one are comparatively higher, because of the involvement of Lys187, Cys156, Pro176, Cys175, Ser179, Gly178, Asn180, and Ser 183 in its periphery (Fig. 3C). The average length of all the sub tunnels of tunnel cluster-1 was 1.495986425 Å, while tunnels cluster-2 and 3 contain 9.098689431 Å and 10.46412388 Å respectively (Fig. 3D). Similarly, the average curvature radiuses of the tunnel clusters were 1.008610157, 1.418848638, and 1.363982938 for cluster-1, 2, and 3 respectively (Fig. 3E). In addition, the average bottleneck radius of the three tunnel clusters of CD81 was 1.870566512, 1.020206265, and 0.998577067 respectively (Fig. 3F).
Pharmacokinetic analysis of the ligands
All four ligands (α-amyrin, β-amyrin, and δ-amyrin as test, and benzyl salicylate as control) showed significant maximum-tolerated doses (MTD), maintaining the range of -0.56 to 0.736 log mg/kg/day. The Amyrin subunits have no hepatotoxicity or AMES toxicity, while the control ligand shows partial hepatotoxicity. There was no violence regarding Lipinski's rules in any of the three test ligands (Table 1). The values of Log P of Amyrins were more than 8.0, while the control's value of Log P was only 2.74. Except for α-amyrin, all three compounds' total clearance (TC) values were positive (< 0.7). The blood–brain-barrier values of the test ligands were over 0.65, almost double that of the control. Among the test ligands, α-amyrin resulted in maximum intestinal absorption (94.062%). The LD50 level of α-amyrin was the highest (2.48) among all the ligands (Table 1). The QSAR profile indicates strong anti-microbial potentialities in α-amyrin than in the others, whereas an anti-infective efficacy in benzyl salicylate has promising anti-infectivity. Surprisingly, all the ligands (including control) showed strong antiviral effects (Table 2).
Supramolecular docking and post-docking analysis
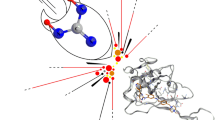
The control ligand benzyl salicylate has the lowest binding affinity (− 6.8 kcal/mol). In contrast, α-amyrin had a comparatively higher binding energy (− 7.3 kcal/mol) than the control ligand. The highest binding energy within the ligand–protein complexes resulted from the β and δ-amyrin (− 7.9 kcal/mol). Considering the binding affinities (Kcal/mol) and the RMSD (Å) values following both the upper and lower bound scores, the amyrin subunits were found superior to the control ligand (Table 3). The control ligand benzyl salicylate forms two hydrogen bonds with 1G8Q macromolecule at Asn184 (3.02 Å) and Thr166 (2.97 Å) amino acids, and six hydrophobic bond interactions with amino acid residues Ile161, Leu185, Lys187, Asp189, Ser160, and Thr 163 (Table 4). The candidate ligand compound α-amyrin formed no hydrogen bond interaction within the ligand-macromolecule complex. However, it exhibits five hydrophobic bond associations on Leu41, Lys26, Leu25, Ile24, and Asn42 residues (Fig. 5A). Similarly, β-amyrin forms one hydrogen bond interaction with Glu16 (3.11 Å) on the CD81 receptor (Fig. 5B) and six hydrophobic interactions with Trp36, Asp15, Asp33, Phe47, Cys32, Glu31 (Table 4). Lastly, the ligand compound δ-amyrin interconnects with Asp23 residue by forming a hydrogen bond length of 2.97 Å (Fig. 5C) and six hydrophobic interactions, namely- Lys20, Asn42, Ile24, Leu41, Glu40, and Lys26 of 1G8Q macromolecule (Table 4).
Molecular dynamic simulation (100 ns)
The RMSD values of the control ligand benzyl salicylate in association with protein ranged between 0 Å to 1.759 Å. The lowest value for calculating the RMSD value is set at 0 Å for all the test ligands. The candidate ligand α-amyrin display the highest RMSD value among the four ligands at 2.053 Å, followed by β-amyrin and δ-amyrin at 1.759 Å and 1.748 Å, respectively (Fig. 6A). On the contrary, the ligand RMSD profile of α-amyrin was distinctive (0 Å to 38.037 Å) compared to the other three candidates including the control Benzyl salicylate. In contrast, β-amyrin took the highest range between (0 Å to 49.768 Å) (Fig. 6B). The interactive alpha carbon atoms (Cα) resulted between 0.345 and 4.02 for the control ligand benzyl salicylate. In contrast, α-amyrin scored the lowest number of α-Carbon interactions ranging between 0.331–3.681. In contrast, beta amyrin and delta amyrin provided a similar range between 0.345 and 4.02 (Fig. 6C). The RMSF values ranged from 0.364 to 4.114 Å for the control ligand, which is the highest among all these four ligands. The α-amyrin showed an RMSF fluctuation range from 0.348 Å to 3.705 Å. In contrast, the beta amyrin and delta amyrin showed similar values with upper and lower bond ranges at 4.114 Å to 0.364 Å and 4.092 to 0.368 Å, respectively (Fig. 6D).
At a probe radius of 1.4 Å, the MolSA values (which is equivalent to the van der Waals surface area of a water molecule) of α-amyrin (159.777–169.075 Å2) were the lowest in the molecular surface area range (Fig. 7A), as compared to the rest of the three candidate ligands including the control. The control ligand benzyl salicylate, along with β and δ-amyrin, demonstrate the highest value range, 375.331–390.034 Å2 (Fig. 7A). The control showed a similar capacity to another candidate, β-amyrin (4.25–4.436 nm) (Fig. 7B). The delta amyrin, provides a slight deviation from the above value of the other two with a range between 4.25 nm and 4.371 nm (Fig. 7B). Following other MDS parameters, α-amyrin exhibited lowest value range distinct from others. In case Rg, which is 2.37–2.814 nm. In contrast, in the control ligand benzyl salicylate, the radius of gyration (Rg) was 4.209–4.43 nm (Fig. 7C). The α-amyrin-1G8Q complex show the highest PSA values (109.106–121.875 Å), which is much higher than the values of control benzyl salicylate, candidate β, and δ-amyrin ranging from 33.324 Å to 41.648 Å (Fig. 7C).
The SASA analysis of the 57 residues of the CD81 macromolecule in association with the four different ligands revealed that at water probe radius 1.4 Å, the polar and apolar energy were 1914.83 Å2 and 2196.92 Å2 (Table 5), respectively for both the β-amyrin and δ-amyrin. However, α-amyrin showed the lowest range of fluctuation profile with a lower value of 104.599 Å2 and an upper value of 352.148 Å2 (Fig. 7D). The control ligand benzyl salicylate, along with β, and δ-amyrin produced the highest fluctuation range (314.694–662.525 Å2) surprisingly (Table 5). Besides, there was no fluctuation in the gradient of calculation, and different unknown areas were observed in every ligand to solvent (water) association (Fig. 7D).
According to the MMGBSA α-dG Binding Score, α-amyrin obtained the highest negative values − 55.00333347 kcal/mol), as followed by β, and δ-amyrin with − 47.37252451 and − 43.88297528 kcal/mol respectively. In contrast, the control scored − 36.28429945 kcal/mol as the endpoint energy (Table 6).
CD81-mediated gene strings behind the protein–protein clustering
The CD81 gene (responsible for encoding CD81 receptor protein) strongly interacts with a group of genes of different functions (Fig. 8). The overall CD81 gene-string network comprises several genes. In contrast, the coexpression of CD81 was observed along with the genes- IFITM1, IFITM3, CD37, C3, CD9, CD19, CD82, CD63, TSPAN4, ITGA4, GFI1, IGSF8, ITGB1, PI4KA, and PTGFRN (Fig. 8A). Particular genes are responsible for entering foreign particles inside the cellular system via coding virus-protein selective receptors were found correlated and interconnected with the CD81 gene, which includes- IFITM1, OFOTM2, and IFITM3 (Fig. 8B). The coded proteins of the interactive genes including C81 were identified and clustered. In protein clustering, proteins like- CLDN1, CD19, CD4, CR2, and IGSF8 were found to be significant for CD81 receptor protein, according to their genetic expression modules. CD19 possessed the second-highest clustering capacities next to CD81 (Fig. 8C).
In vivo assessments
Considering all the parameters considered for the in vivo study, β-amyrin was the most effective targeted compound in protecting against HCV viral infection and reducing the risks of hepatic fibroblasts. As compared to the control (CG) and the negative control (NCG) groups, the treatment group 1 (TG-1) and 2 (TG-2) got significant results (P < 0.0001), where doses effects played a pivotal role in all aspects in determining the TNF-α (Fig. 9). The impact of using DMN in combination with the β-amyrin results is almost equal to the group administrated DMN-free supplements of β-amyrin means P < 0.001 in all the cases (Tables 7, and 8). Due to having most values in the range of standard pharmacological parameters, β-amyrin was found best among the others in vivo because α-amyrin fluctuates the basic serological parameters comparatively more randomly than β.
Discussion
HCV infection affects roughly 210 million people worldwide, with an average prevalence of 3% (Jafri and Gordon 2018) of the global population, demonstrating the need for effective drugs and therapeutics. However, multiple vaccine candidates have gone through the development process (Ansaldi et al. 2014). Phytocompounds generated from plants have physiologically active qualities, such as antiviral, anti-inflammatory, antifungal, and antibacterial activities, with partial adverse effects. HCV resistance has been found in several phytocompounds (Patil et al. 2022), among which both α and β amyrin can exhibit hyperactivity in silico pharmacokinetics. This makes them suitable HCV candidate therapeutics (Zhang et al. 2022).
Active-site identification considering quantum tunneling of CD81 protein
The active sites of the CD81 receptor were predicted to understand the best active binding sites of the protein (Fig. 2) following the qualitative and quantitative measurements using CASTp and COACH-D algorithms, respectively (Nipun et al. 2021). Among the active sites, the fourth one was found more viable (Fig. 2D), considering different predicted amino acid residues, binding energy, and binding strength obtained (Fig. 2E).
Quantum tunnel properties like- tunnel length, curvature, and bottleneck radius (Nipun et al. 2021), validate the potential of accommodating the test ligands with the best possible binding affinities (Fig. 3). In this in-silico study, three significant tunnels are found in the study protein receptor CD81, where each of the tunnels contains seven sub-tunnel clusters. Each sub-tunnel is unique and distinctive, established on the number of amino acid residues at their bottleneck point. From this qualitative analysis of these tunnels, it is prominent that the third major tunnel has a higher length and curvature involved with more amino acids than the other two tunnels (Fig. 3A–C). The average length of the third tunnel cluster was highly significant (10.46412388 Å) than cluster-1 and 2 (1.495986425 Å and 9.098689431 Å), respectively (Fig. 3D). In contrast, the second tunnel cluster possesses the best curvature radius (1.418848638) among all the clusters (Fig. 3E). Additionally, the bottleneck radius of the first tunnel cluster was the highest (1.870566512) of any other clusters identified (Fig. 3F). All the parameters above mean- tunnel length, curvature, and bottleneck radius are the major factors to validate the predictive super docking position of any protein's active site (Kingsley and Lill 2014).
Profiling of the in silico pharmacokinetics of the ligands
In the ADMET analysis, according to Lipinski's rules, five selective parameters were analyzed, which include a molecular weight of 500; a maximum of five hydrogen bond donors; a maximum of 10 hydrogen bond acceptors; a partition coefficient (log P) of 5; and a molar refractivity of 40–130 (Mane et al. 2022). The molecular weight for all our ligands was less than 500. However, Log P for only the benzyl salicylate (control ligand) was less than five, and the rest (test ligands) were more than 8. Hydrogen bond donors were 1 for all the drugs, and hydrogen bond acceptors were 3 for benzyl salicylate and 1 for the other drugs. Overall, there was no violence related to Lipinski's rules for any targeted ligands (Table 1). A QSAR analysis was conducted to ensure the accuracy of the major compounds (Parikesit et al. 2021). According to the QSAR profile, α-amyrin was identified as an antimicrobial agent, while anti-infective activity was only found in benzyl salicylate (Table 1). In addition, antiviral potentialities were seen in ale four ligands, which validates the selection of the lead ligand compounds for targeting CD81 (Table 2).
Supramolecular docking and post molecular docking analysis of the superpositions' of ligands
The supramolecular docking of the control ligand (Fig. 4) and the test ligands (Fig. 5) at the most active site of the CD81 receptor revealed diversified binding affinities (Kcal/mol) following their upper-bound and lower-bound RMSD (Å) values (Table 3). Depending on the molecular docking scores of the parameters (Paul et al. 2022), in the present study- β and δ-amyrins showed their significant potentiality over the others with − 7.9 kcal/mol energy (Table 3). The interactions of different atoms of the ligands with the amino acid residues of CD81 were observed following the nature of the hydrogen bonds and hydrophobic interactions among them [34,36]. The control ligand benzyl salicylate formed two hydrogen bonds (Fig. 4B, C), whereas the β and δ-amyrins formed a single bond each (Fig. 5B, C). Though α-amyrin didn't form any hydrogen bond specifically (Fig. 5A), it possessed five different hydrophobic interactions with the neighboring amino acid residues of CD81 (Table 4). Usually, both hydrogen bond and hydrophobic interactions (non-covalent) are needed to profile the therapeutic efficiencies of the test ligands, where the increased amount of hydrogen bonds stands for the increased targeted-binding strength of the ligands of interest (Paul et al. 2022). Depending on the binding affinity (Kcal/mol), the control ligand is inferior to all the ligands with -6.8 kcal/mol energy, despite forming a single hydrogen bond (Table 3).
The complex formed between the control ligand benzyl salicylate and the CD81 receptor (A), where the qualitative ligand–protein interaction is represented (B), along with their quantitative measurements considering the hydrogen bond interactions and non-covalent (hydrophobic) interactions (C). Hydrogen bonds are referred to as green lines, whereas the red lines represent hydrophobic (noncovalent interactions)
Representation of the hydrogen bonds and hydrophobic interactions of the ligand atoms with different amino acid residues at the point of their supramolecular docking. Though α-amyrin formed no hydrogen bonds (A), β-amyrin formed a single hydrogen bond with Glu16 (3.11 Å) (B); whereas δ-amyrin formed with Asp23 (2.97 Å) (C)
Molecular dynamic simulation (100 ns)
The molecular dynamic simulation (MDS) expresses the behavior of proteins in detail when in complex with the targeted ligands for a certain period (ns) (Hollingsworth and Dror 2018). It provides a perspicuous idea of the stability of the protein–ligand complexes (Jamroz et al. 2013). In the current study, the MDS of the ligand-free1 was initially conducted for 10 ns on CABS-flex 2.0 to observe natural changes in structural orientation and interactions with adjacent water molecules and ions (Kurcinski et al. 2015). The ligand–protein interactions were observed for 3.1 ns using LARMD (Yang et al. 2020]. Finally, 100 ns of the molecular dynamic simulation was conducted comprehensively with Desmond-Schrodinger to determine the parameters (Yang et al. 2020; Ivanova et al. 2018) root mean square deviation (RMSD), root means square fluctuation (RMSF), the radius of gyration (Rg), solvent-accessible surface area (SASA), polar surface area (PSA), and molecular surface area (MolSA), MMGBSA dG scores (Figs. 6 and 7). The RMSD values of the candidate ligands were within a justifiable range observed (Fig. 6A), where the minimum values didn't cross 3 Å. Lower RMSD values indicate more stable conformation of ligands, intensely observed for β and δ-amyrins, ensuring that enzymatic structure is not hampered. In the beginning, RMSD values for all the ligands were 0 Å, respectively, and α-amyrin displayed the highest RMSD value, 2.053 Å, after 100 ns of MDS. However, β and δ-amyrins demonstrated lower RMSD values at 1.759 Å each (Fig. 6A). The control ligand Benzyl-salicylate scored between 0 Å to 1.759 Å as reported in established literature as well (Olaby et al. 2013). In the case of the ligand RMSD, the same phenomena were found for both the β and δ-amyrins, where the α-amyrin fluctuation range was the highest, making it a less effective drug candidate (Fig. 6B) than others.
The Cα values were significant for the β and δ-amyrins 0.45 to 3.88 at the end of the 100 ns molecular dynamic simulation. In contrast, the values for α-amyrin were scored from 0.39 to 3.68, making itself a comparatively less stable ligand candidate against the CD81 receptor (Fig. 6C). In RMSF calculation, high divergence and flexibility during a simulation indicate that the protein structure is weak and unstable (Ghosh et al. 2021). RMSF profiles of all protein–ligand complexes from 57 protein residues underwent crucial alterations over time. The control ligand exhibits RMSF values from 0.364 to 4.114 Å, the highest among all the four ligand–protein complexes. The α-amyrin shows RMSF fluctuation differences from 0.348 Å to 3.705 Å, in contrast to β and δ-amyrin. Here they exhibit similar values with an upper bound of 4.114 Å and a lower bound of 0.364 Å (Fig. 6D). Similar findings have been reported in several recent pieces of literature (Dutta et al. 2021).
The molecular surface area (MolSA) represents the stability profiles of any ligands-protein complex in MDS (Arefin et al. 2021). After the 100 ns molecular simulation, the α-1G8Q complex demonstrated the lowest value and was thus considered the least stable ligand (Fig. 7A). On the contrary, the rest of the three ligands (benzyl salicylate, β, and δ-amyrins) formed stable complexes according to their MolSA values. Similarly, the radius of gyration (Rg) is another vital parameter for evaluating the stability of the docked protein–ligand complexes, which measures the distance between the center of mass and the terminal protein (Yamamoto et al. 2021). Due to having the lowest radius of gyration values, α-amyrin was found to be comparatively stable. The higher Rg value represents fragile protein conformation, whereas the lower Rg value depicts the compactness of the protein molecule (Zaki et al. 2022). In this sense, the higher Rg values of the control ligand, β, and δ-amyrins, revealed their weak protein folding rendering and compactness (Fig. 7B). This description of Rg values is strongly supported by other literature (Ayyamperumal et al. 2021).
Polar surface area is one of MDS's key elements, which deals with the accessibility of the drug molecule inside the blood–brain barrier (Alsenan et al. 2020). The accepted range of PSA of a proper drug molecule exhibiting the ability to cross the blood–brain barrier falls between 40 Å2 < X ≤ 90 Å2 (X = substance) (Nipun et al. 2021). In this current study, the α-amyrin ligand exemplifies a much higher PSA value ranging from 109.106 Å2 to 121.875 Å2, deviating from the standard range. The rest of the ligands, including the control, were within an inclusive PSA range of 33.314 Å2 < X ≤ 41.648 Å2 (X = substance). It is a very positive result for the β and δ-amyrins to be used as potential drugs for the targeted site of CD81 (Fig. 7C). On the other hand, the drug binding to a protein can influence the solvent-accessible surface area. Hydrophobic interaction is one of the essential intermolecular interactions between the docked protein–ligand complex and the surrounding water molecules. Hydrophobic bonds are established among non-polar amino acids to ensure the stability of the protein–ligand complex in an aqueous environment solution (Shi et al. 2019). The shielding of the non-polar amino acids in hydrophobic cores moves away from the aqueous environment leading to decreased SASA values. In this in silico study, after the 100 ns molecular dynamic simulation, only α-amyrin provided the lowest SASA value range (104.599–352.148 Å2) than the other ligands, including the control (Fig. 7D), referring to α-amyrin as the superior ligand to others (Khan et al. 2020). The total area-to-energy ratio for all the ligands, based on the water probe radius (1.4 Å), was the same for both the polar (1914.83) and apolar (2196.92) considerations. No unknown surface was available for all the ligand–protein complexes (Table 5). According to the MMGBSA dG binding score, all the amyrin subunits showed an extreme endpoint energy level (Kcal/mol) than that of the control ligand (Table 6), even at the very last frames of the molecular dynamics simulation. A higher negative value could be achieved, and the more stable ligand–protein complex formation is considered an essential precursor for in silico bioprospecting of drug candidates (Pandey et al. 2015).
Analysis of the gene–gene interaction (GGI) behind the protein–protein interaction (PPI) of CD81
The CD81 receptor protein contains a very complex string networking in its genetic stage from which the signal of clustering with other related proteins is triggered. The CD81 gene is the main coder of the CD81 receptor, which interacts with a group of genes. Among all those genes, the relationship among the CD81, CD82, CD19, and IFITM1 was found to be highly significant (Wilkins et al. 2013; Narayana et al. 2015; Harris et al. 2010) in terms of genetic coexpression inside the living systems (Fig. 8A). Unlike any average coexpression, not all genes in any gene-string network are essential for allowing the organism to insert any foreign particle (virus) inside its cells. Instead, there are mainly corresponding genes whose compromised function paves the way for influxing any viral particle (E2 protein of HCV in this case) inside the cell. In case of HCV infection, the E2 receptor is required to take the help of several interactive genes from the exact string, such as CD81, IFITM1, IFITM2, and IFITM3 (Fig. 8B). The genetic signaling and coexpression of the CD81 receptor protein, starts to follow clustering with the related proteins- IGSF8, CLDN1, CR2, CD4, and CD19. The clustering of CD4 and CD19 with CD81 protein is rigorous (Fig. 8C). This phenomenon is common in HCV research in human cellular systems (Narayana et al. 2015; Harris et al. 2010).
CD81 mediated overall genetic string interactions among the coexpressed genes are represented (A) simultaneously with the gene clusters responsible for regulating HCV E2 protein entry inside the cell upon any compromization (B). In addition, the genetic information of the string of CD81 gene in establishing CD81 receptor protein clusters with the other protein groups responsible for protecting cells from E2 protein attachment was established (C). The gold-colored genes (B) are the cluster of protecting cells from the HCV influx mechanism
Alike phytochemical sources, most often selective functional bioactive components like fatty acids, antimicrobial peptides, and secondary therapeutic derivatives are derived from different microorganisms (Dipta et al. 2021a, b; Azad et al. 2016), where many of which are clinically considered probiotics (Abdullah-Al-Mamun et al. 2016). These bioactive compounds can render proper opsonization as part of our secondary immune response against infectious diseases including HCV infection and cancer (Al Azad et al. 2016; Al Azad and Khan 2019).
In vivo analysis of the liver-specific serological parameters
In this current study, β-amyrin consolidated its potentialities as a target-specific novel compound, resulting in significant scores in basic serological and HCV-specific serological parameters as P < 0.0001 for TNF-α (Fig. 9) and P < 0.001 (Tables 7, and 8) respectively in the scale of significance α = 0.05. Among the HCV and consequent hepatic fibrosis-specific parameters, the secretion and co-existence of different liver enzymes were estimated, such as ALP (IU/L) (alkaline phosphatase), LDH (IU/L) (lactate dehydrogenase), AST (IU/L) (aspartate aminotransferase), ALT (IU/L) (alanine transaminase) (Table 8). The values obtained from the experimental groups' post-trial analysis were almost similar to the previously reported findings (Krishnan et al. 2014; Parvez et al. 2018; Thirupathi et al. 2017). The results from the group administrated β-amyrin with and without DMN resulted very closely. The concentrations of MDA (nmol/ml) among the OSM and antioxidant parameters were maximum in volume, valuing 11.89 ± 0.87 (nmol/ml) (TG-1) and 12.02 ± 0.65 (nmol/ml) (TG-2) as compared to 14.26 ± 0.21 (nmol/ml) (CG), and 20.13 ± 0.86 (nmol/ml) (NCG) (Table 7). Similar to these, LDH was the highest in volume among the liver-specific enzymes secreted during HCV and hepatic fibroblast mean 76.43 ± 3.09 (IU/L) (TG-1) and 79.23 ± 2.54 (IU/L) (TG-2) as compared to 115.14 ± 3.11 (IU/L) (NCG), ensuring P < 0.001 (Table 8). These values strongly matched the research outputs of some previous studies, including Parvez et al. (2018), Sharif et al. (2021), and Thirupathi et al. (2017). Reactive oxygen species (ROS) has become an excellent factor in determining the antiviral and anticancer activity of different phytochemical pharmacophores where β-amyrin can be a very effective target specific therapeutic similar to the quercetin, studied previously (Biswas et al. 2022).
Limitations of this study
In some frames of the molecular dynamic simulation, the SASA values fluctuated randomly with the increase of temperature issues sometimes that were overcome instantly in the following steps. Though α-amyrin showed very strong ADMET, QSAR, and MDS properties, it surprisingly failed to form a hydrogen bond in the active site of the CD81 protein. The docking process was repeated several times between α-amyrin and CD81, but the same result was found each time, which can be considered another finding. Besides, comprehensive serological and immunological assessment approaches are needed to analyze the in vivo feasibility of using those three subunits of amyrin as target-specific drugs. Though the authors' are working on that as well, the work will require much time to be accomplished.
Conclusion
Considering the frequent infection and fatality of HCV, the current study aimed to figure out the target specificity and therapeutic efficacy of the three different subunits of amyrin from the list of 203 potential bioactive compounds. Diversified in silico pharmacokinetic and pharmacodynamic parameters were analyzed. Afterward, the molecular dynamic simulation was conducted comprehensively for 100 ns to reveal the target-specificity of the β and δ-amyrin on the superactive position of the CD81 receptor, identified through the quantum tunneling algorithm. Values from the parameters mean- RMSD, RMSF, Rg, SASA, Cα, MolSA, and PSA resulted within the favorable ranges for all amyrin subunits when complexed. Finally, CD81-mediated gene clusters were identified to know about responsible ones for regulating the protein–protein infection (PPI) pathways during the HCV infection inside the cells. Following the in silico pharmacokinetic, pharmacodynamic, and molecular-string profiles, the present research strongly recommends the amyrin subunits to be used as prophylactics for HCV infection in humans as a targeted drug on the CD81 receptor protein. Despite the full-length in silico bioprospecting, the amyrins have been taken for further in vivo analysis in continuation to this study to validate the doses in the mice model, and the in vivo research is still underway.
Data availability
All the data will be available from the Corresponding Author upon reasonable request from the journal authority.
References
Abdullah-Al-Mamun M, Jakir Hasan M, Al Azad S et al (2016) Evaluation of potential probiotic characteristics of isolated lactic acid bacteria from goat milk. Biotechnol J Int 14(2):1–7. https://doi.org/10.9734/BBJ/2016/26397
Akter KM, Tushi T, Jahan Mily S, Mohona RA, Anis S, Chakraborty AK, Tabassum E, Islam TU, Akhi OJ, Nishe IS, Laxy BN, Zerin SS, Roble AJ, Hossain MI, Ahmed S, Azad SA (2020) RT-PCR mediated identification of SARS-CoV-2 patients from particular regions of Bangladesh and the multi-factorial analysis considering their pre and post infection health conditions. Biotechnol J Int 24(6):43–56. https://doi.org/10.9734/bji/2020/v24i630121
Al Azad S, Khan I (2019) HAMLET (Human Alpha Lactalbumin Made Lethal to Tumor Cells)-a hope for the cancer patients. Adv Pharmacol Clin Trials 4(1):000152. https://doi.org/10.23880/apct-16000152
Al Azad S, Shahriyar S, Mondal KJ (2016) Opsonin and its mechanism of action in secondary immune. J Mol Stud Med Res 1(02):48–56. https://doi.org/10.18801/jmsmr.010216.06
Al Azad S, Ahmed S, Biswas P (2022) Quantitative analysis of the factors influencing IDA and TSH downregulation in correlation to the fluctuation of activated vitamin D3 in women. J Adv Biotechnol Exp Ther 5(2):320–333. https://doi.org/10.5455/jabet.2022.d118
Alsenan S, Al-Turaiki I, Hafez A (2020) A recurrent neural network model to predict blood–brain barrier permeability. Comput Biol Chem 89:1077
Ansaldi F, Orsi A, Sticchi L, Bruzzone B, Icardi G (2014) Hepatitis C virus in the new era: perspectives in epidemiology, prevention, diagnostics and predictors of response to therapy. World J Gastroenterol 20(29):9633
Arefin A, Ismail Ema T, Islam T, Hossen S, Islam T, Al Azad S, Uddin Badal N, Islam A, Biswas P, Alam NU, Islam E, Anjum M, Masud A, Kamran S, Rahman A, Kumar PP (2021) Target specificity of selective bioactive compounds in blocking α-dystroglycan receptor to suppress Lassa virus infection: an insilico approach. J Biomed Res 35(6):459–473. https://doi.org/10.7555/JBR.35.20210111
Asselah T, Bieche I, Sabbagh A, Bedossa P, Moreau R, Valla D, Vidaud M, Marcellin P (2009) Gene expression and hepatitis C virus infection. Gut 58(6):846–858
Ayyamperumal S, Dhananjay DJ, Tallapaneni V, Mohan S, Basappa S, Selvaraj J, Joghee NM, Chandrasekar MJ (2021) Molecular docking analysis of α-Topoisomerase II with δ-Carboline derivatives as potential anticancer agents. Bioinformation 17(1):249
Azad SA, Mamun MAA, Mondal KJ, Alim S, Rahman MM (2016) Range of various fungal infections to local and hybrid varieties of non-germinated lentil seed in Bangladesh. J Biosci Agric Res 09(01):775–781. https://doi.org/10.18801/jbar.090116.93
Azad SA, Moazzem Hossain K, Rahman SMM, Al Mazid MF, Barai P, Gazi MS (2020a) In ovo inoculation of duck embryos with different strains of Bacillus cereus to analyse their synergistic post-hatch anti-allergic potentialities. Vet Med Sci 6(4):992–999. https://doi.org/10.1002/vms3.279
Azad SA, Farjana M, Mazumder B, Abdullah-Al-Mamun M, Haque A (2020b) Molecular identification of a Bacillus cereus strain from Murrah buffalo milk showed in vitro bioremediation properties on selective heavy metals. J Adv Vet Anim Res 7(1):62–68. https://doi.org/10.5455/javar.2020.g394
Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS (2016) Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44(W1):W147–W153
Biswas P, Dey D, Biswas PK, Rahaman TI, Saha S, Parvez A, Khan DA, Lily NJ, Saha K, Sohel M, Hasan MM, Al Azad S, Bibi S, Hasan MN, Rahmatullah M, Chun J, Rahman MA, Kim B (2022) A comprehensive analysis and anticancer activities of quercetin in ROS-mediated cancer and cancer stem cells. Int J Mol Sci 23(19):11746. https://doi.org/10.3390/ijms231911746
Brandt BW, Heringa J, Leunissen JA (2008) SEQATOMS: a web tool for identifying missing regions in PDB in sequence context. Nucleic Acids Res 36(suppl_2):W255–W259
Bruening J, Lasswitz L, Banse P, Kahl S, Marinach C, Vondran FW, Kaderali L, Silvie O, Pietschmann T, Meissner F, Gerold G (2018) Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB. PLoS Pathog 14(7):e1007111
Burlone ME, Budkowska A (2009) Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J Gen Virol 90(5):1055–1070
Chen SL, Morgan TR (2006) The natural history of hepatitis C virus (HCV) infection. Int J Med Sci 3(2):47
Chigbu DI, Loonawat R, Sehgal M, Patel D, Jain P (2019) Hepatitis C virus infection: host–virus interaction and mechanisms of viral persistence. Cells 8(4):376
Dipta D, Paul PK, Al Azad S, Al Mazid MF, Khan AM, Sharif MA, Rahman MH (2021a) Molecular optimization, docking, and dynamic simulation profiling of selective aromatic phytochemical ligands in blocking the SARS-CoV-2 S protein attachment to ACE2 receptor: an in silico approach of targeted drug designing. J Adv Vet Anim Res 8(1):24–35. https://doi.org/10.5455/javar.2021.h481
Dipta D, Ema TI, Biswas P, Aktar S, Islam S, Rinik UR, Firoz M, Ahmed SZ, Azad SA, Rahman A, Afrin S, Mahedi RA, Md. Nasir UB (2021b) Antiviral effects of bacteriocin against animal-to-human transmittable mutated SARS-COV-2: a systematic review. Front Agric Sci Eng 8(4):603–622. https://doi.org/10.15302/J-FASE-2021397
Durán-Iturbide NA, Díaz-Eufracio BI, Medina-Franco JL (2020) In silico ADME/Tox profiling of natural products: a focus on BIOFACQUIM. ACS Omega 5(26):16076–16084
Dutta T, Baildya N, Khan AA, Ghosh NN (2021) Inhibitory effect of anti-HIV compounds extracted from Indian medicinal plants to retard the replication and transcription process of SARS-CoV-2: an insight from molecular docking and MD-simulation studies. Netw Model Anal Health Inform Bioinform 10(1):1–1
Farquhar MJ, Harris HJ, McKeating JA (2011) Hepatitis C virus entry and the tetraspanin CD81. Biochem Soc Trans 39(2):532–536
Fénéant L, Levy S, Cocquerel L (2014) CD81 and hepatitis C virus (HCV) infection. Viruses 6(2):535–572
Ferdausi N, Islam S, Rimti FH, Quayum ST, Arshad EM, Ibnat A, Islam T, Arefin A, Ema TI, Biswas P, Dey D, Azad SA (2022) Point-specific interactions of isovitexin with the neighboring amino acid residues of the hACE2 receptor as a targeted therapeutic agent in suppressing the SARS-CoV-2 influx mechanism. J Adv Vet Anim Res 9(2):230–240. https://doi.org/10.5455/javar.2022.i588
Ghosh R, Chakraborty A, Biswas A, Chowdhuri S (2021) Evaluation of green tea phenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn 39(12):4362–4374
Grebely J, Prins M, Hellard M, Cox AL, Osburn WO, Lauer G, Page K, Lloyd AR, Dore GJ (2012) Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. Lancet Infect Dis 12(5):408–414
Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H, Balfe P (2010) Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem 285(27):21092–21102
Hollingsworth SA, Dror RO (2018) Molecular dynamics simulation for all. Neuron 99(6):1129–1143
Islam R, Akter KM, Rahman A, Khanam NN, Al Azad S, Islam MR, Farjana M, Rahman MH, Badal MN, Ahmed S (2021) The serological basis of the correlation between iron deficiency anemia and thyroid disorders in women: a community based study. J Pharm Res Int 30(19A):69–81. https://doi.org/10.9734/jpri/2021/v33i19A31330
Ivanova L, Tammiku-Taul J, García-Sosa AT, Sidorova Y, Saarma M, Karelson M (2018) Molecular dynamics simulations of the interactions between glial cell line-derived neurotrophic factor family receptor GFRα1 and small-molecule ligands. ACS Omega 3(9):11407–11414
Jafri SM, Gordon SC (2018) Epidemiology of hepatitis C. Clin Liver Dis 12(5):140
Jamroz M, Kolinski A, Kmiecik S (2013) CABS-flex: server for fast simulation of protein structure fluctuations. Nucleic Acids Res 41(W1):W427–W431
Jardim AC, Shimizu JF, Rahal P, Harris M (2018) Plant-derived antivirals against hepatitis c virus infection. Virol J 15(1):1–3
John A, Umashankar V, Krishnakumar S, Deepa PR (2015) Comparative modeling and molecular dynamics simulation of substrate binding in human fatty acid synthase: enoyl reductase and β-Ketoacyl reductase catalytic domains. Genom Inf 13(1):15–24. https://doi.org/10.5808/GI.2015.13.1.15
Kalinina OV (2015) Hepatitis C virus: variability mechanisms, classification, evolution. Probl Virol 60(5):5–10
Khan A, Khan M, Saleem S, Babar Z, Ali A, Khan AA, Sardar Z, Hamayun F, Ali SS, Wei DQ (2020) Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscip Sci 12(3):335–348
Kingsley LJ, Lill MA (2014) Ensemble generation and the influence of protein flexibility on geometric tunnel prediction in cytochrome P450 enzymes. PLoS ONE 9(6):e99408. https://doi.org/10.1371/journal.pone.0099408
Krishnan K, Mathew LE, Vijayalakshmi NR, Helen A (2014) Anti-inflammatory potential of β-amyrin, a triterpenoid isolated from Costus igneus. Inflammopharmacology 22(6):373–385. https://doi.org/10.1007/s10787-014-0218-8
Kumar A, Singh AK, Tripathi G (2020) Phytochemicals as potential curative agents against viral infection: a review. Curr Org Chem 24(20):2356–2366
Kurcinski M, Jamroz M, Blaszczyk M, Kolinski A, Kmiecik S (2015) CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res 43(W1):W419–W424
Kuriata A, Gierut AM, Oleniecki T, Ciemny MP, Kolinski A, Kurcinski M, Kmiecik S (2018) CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures. Nucleic Acids Res 46(W1):W338–W343. https://doi.org/10.1093/nar/gky356
Lavanchy D (2011) Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17(2):107–115
Lavie M, Sarrazin S, Montserret R, Descamps V, Baumert TF, Duverlie G, Séron K, Penin F, Dubuisson J (2014) Identification of conserved residues in hepatitis C virus envelope glycoprotein E2 that modulate virus dependence on CD81 and SRB1 entry factors. J Virol 88(18):10584–10597
Levy S, Todd SC, Maecker HT (1998) CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol 16:89
Li M, Li D, Tang Y, Wu F, Wang J (2017) CytoCluster: a cytoscape plugin for cluster analysis and visualization of biological networks. Int J Mol Sci 18(9):1880. https://doi.org/10.3390/ijms18091880
Li HC, Yang CH, Lo SY (2021) Cellular factors involved in the hepatitis C virus life cycle. World J Gastroenterol 27(28):4555
Lindenbach BD, Rice CM (2005) Unravelling hepatitis C virus replication from genome to function. Nature 436(7053):933–938
Lyne PD, Lamb ML, Saeh JC (2006) Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J Med Chem 49(16):4805–4808
Mane A, Kasibhatla SM, Vidhate P, Saxena V, Patil S, Rao A, Nirmalkar A, Kulkarni-Kale U, Panda S (2022) Phylogenetic analysis of spread of hepatitis C virus identified during HIV outbreak investigation, Unnao India. Emerg Infect Dis 28(4):725
Millman AJ, Nelson NP, Vellozzi C (2017) Hepatitis C: review of the epidemiology, clinical care, and continued challenges in the direct-acting antiviral era. Curr Epidemiol Rep 4(2):174–185
Molinari MD, Mendon CAJ, Barbosaa D, Mrin D, Mertz-Henning LM, Nepomuceno A (2021) Transcriptome analysis using RNA-Seq from experiments with and without biological replicates: a review. Revista Ciência Agrária 64:1–13
Moradpour D, Penin F, Rice CM (2007) Replication of hepatitis C virus. Nat Rev Microbiol 5(6):453–463
Morozov VA, Lagaye S (2018) Hepatitis C virus: morphogenesis, infection and therapy. World J Hepatol 10(2):186
Morshed AKMH, Al Azad S, Mia MAR et al (2022) Oncoinformatic screening of the gene clusters involved in the HER2-positive breast cancer formation along with the in silico pharmacodynamic profiling of selective long-chain omega-3 fatty acids as the metastatic antagonists. Mol Divers. https://doi.org/10.1007/s11030-022-10573-8
Narayana SK, Helbig KJ, McCartney EM, Eyre NS, Bull RA, Eltahla A, Lloyd AR, Beard MR (2015) The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J Biol Chem 290(43):25946–25959
Nipun TS, Ema TI, Mia MAR, Hossen MS, Arshe FA, Ahmed SZ, Masud A, Taheya FF, Khan AA, Haque F, Azad SA, Al Hasibuzzaman M, Tanbir M, Anis S, Akter S, Mily SJ, Dey D (2021) Active site-specific quantum tunneling of hACE2 receptor to assess its complexing poses with selective bioactive compounds in co-suppressing SARS-CoV-2 influx and subsequent cardiac injury. J Adv Vet Anim Res 8(4):540–556. https://doi.org/10.5455/javar.2021.h544
Oh KK, Adnan M, Ju I, Cho DH (2021) Anetwork pharmacology study on main chemical compounds from Hibiscus cannabinus L. leaves. RSC Adv 11(19):11062–11082. https://doi.org/10.1039/D0RA10932K
Olaby RA, Azzazy HM, Harris R, Chromy B, Vielmetter J, Balhorn R (2013) Identification of ligands that target the HCV-E2 binding site on CD81. J Comput-Aided Mol Des 27(4):337–346
Pandey RK, Sharma D, Bhatt TK, Sundar S, Prajapati VK (2015) Developing imidazole analogues as potential inhibitor for Leishmania donovani trypanothione reductase: virtual screening, molecular docking, dynamics and ADMET approach. J Biomol Struct Dyn 33(12):2541–2553
Parikesit AA, Nurdiansyah R, Garrido G (2021) Natural products repurposing of the H5N1-based lead compounds for the most fit inhibitors against 3C-like protease of SARS-CoV-2. J Pharm Pharmacogn Res 9(5):730–745
Parvez MK, Alam P, Arbab AH, Al-Dosari MS, Alhowiriny TA, Alqasoumi SI (2018) Analysis of antioxidative and antiviral biomarkers β-amyrin, β-sitosterol, lupeol, ursolic acid in Guiera senegalensis leaves extract by validated HPTLC methods. Saudi Pharm J 26(5):685–693
Patel B, Sharma S, Nair N, Majeed J, Goyal RK, Dhobi M (2021) Therapeutic opportunities of edible antiviral plants for COVID-19. Mol Cell Biochem 476(6):2345–2364
Patil VS, Harish DR, Vetrivel U, Roy S, Deshpande SH, Hegde HV (2022) Hepatitis C virus NS3/4A inhibition and host immunomodulation by Tannins from Terminalia chebula: a structural perspective. Molecules 27(3):1076
Paul PK, Al Azad S, Rahman MH, Farjana M, Uddin MR, Dey D, Mahmud S, Ema TI, Biswas P, Anjum M, Akhi OJ, Ahmed SZ (2022) Catabolic profiling of selective enzymes in the saccharification of non-food lignocellulose parts of biomass into functional edible sugars and bioenergy: An in silico bioprospecting. J Adv Vet Anim Res 9(1):19–32. https://doi.org/10.5455/javar.2022.i565
Pfaender S, Brown RJ, Pietschmann T, Steunmann E (2014) Natural reservoirs for homologs of hepatitis C virus. Emerg Microbes Infect 3(1):1–9
Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, De Jong YP, Rice CM (2009) Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457(7231):882–886
Prentoe J, Velázquez-Moctezuma R, Foung SK, Law M, Bukh J (2016) Hypervariable region 1 shielding of hepatitis C virus is a main contributor to genotypic differences in neutralization sensitivity. Hepatology 64(6):1881–1892
Rashaduzzaman M, Kamrujjaman M, Islam MA, Ahmed S, Al Azad S (2019) An experimental analysis of different point specific musculoskeletal pain among selected adolescent-club cricketers in Dhaka City. Eur J Clin Exp Med 17(4):308–314. https://doi.org/10.15584/ejcem.2019.4.4
Sharif MA, Hossen MS, Shaikat MM, Mashuk FA, Haidary TI, Dey DI, Paul PK, Al Azad SA, Al Mazid MF, Badal MN (2021) Molecular optimization, docking and dynamic simulation study of selective natural aromatic components to block E2-CD81 complex formation in predating protease inhibitor resistant HCV influx. Int J Pharm Res. https://doi.org/10.31838/ijpr/2021.13.02.408
Shi X, Wang Y, Sun H, Chen Y, Zhang X, Xu J, Zhai G (2019) Heparin-reduced graphene oxide nanocomposites for curcumin delivery: in vitro, in vivo and molecular dynamics simulation study. Biomater Sci 7(3):1011–1027
Thirupathi A, Silveira PC, Nesi RT, Pinho RA (2017) β-Amyrin, a pentacyclic triterpene, exhibits anti-fibrotic, anti-inflammatory, and anti-apoptotic effects on dimethyl nitrosamine–induced hepatic fibrosis in male rats. Hum Exp Toxicol 36(2):113–122
Tian W, Chen C, Lei X, Zhao J, Liang J (2018) CASTp 3.0: computed atlas of surface topography of proteins. Nucl Acids Res 46(W1):W363–W367. https://doi.org/10.1093/nar/gky473
Warde DF, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38(suppl2):W214–W220
Wilkins C, Woodward J, Lau DT, Barnes A, Joyce M, McFarlane N, McKeating JA, Tyrrell DL, Gale M Jr (2013) IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 57(2):461–469
Wu J, Liu W, Gong P (2015) A structural overview of RNA-dependent RNA polymerases from the Flaviviridae family. Int J Mol Sci 16(6):12943–12957
Yamamoto H, Nakanozo H, Narukawa T (2021) Development of a vibration isolator using air suspensions with slit restrictions. Vibration engineering for a sustainable future. Springer, Cham, pp 271–277
Yang J, Roy A, Zhang Y (2013) Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29(20):2588–2595. https://doi.org/10.1093/bioinformatics/btt447
Yang JF, Wang F, Chen YZ, Hao GF, Yang GF (2020) LARMD: integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief Bioinform 21(6):2206–2218. https://doi.org/10.1093/bib/bbz141
Zaki AA, Ashour A, Elhady SS, Darwish KM, Al-Karmalawy AA (2022) Calendulaglycoside A showing potential activity against SARS-CoV-2 main protease: Molecular docking, molecular dynamics, and SAR studies. J Tradit Complement Med 12(1):16–34
Zaltron S, Spinetti A, Biasi L, Baiguera C, Castelli F (2012) Chronic HCV infection: epidemiological and clinical relevance. BMC Infect Dis 12(2):1–7
Zhang Y, Yang Y, Yan C, Li J, Zhang P, Liu R, He J, Chang YX (2022) A review of the ethnopharmacology, phytochemistry and pharmacology of Cynanchum atratum. J Ethnopharmacol 284:114748
Zona L, Tawar RG, Zeisel MB, Schuster C, Lupberger J, Baumert TF (2014) CD81-receptor associations—impact for hepatitis C virus entry and antiviral therapies. Viruses 6(2):875–892
Acknowledgements
The authors must extend their respect and satisfaction to RPG Organization (Govt. Registration ID: 05-060-06021) for ensuring all technical support in cordial collaboration as part of the excellent organizational technology transfer and faculty exchange program under the Category H2 (ID. #16-2021/22)'.
Funding
No funding was received for this research.
Author information
Authors and Affiliations
Contributions
Conceptualization, methodology, and supervision: MJ; Project administration & co-supervision: SAA; Resources: AJ, MFU, AR; Data curation: SAA, FT, Mst.UH; Writing original draft: PS, AKMHM, SR, FFR, MRS, AHO, TI; Visualization: RT, MS, MJ; Investigation: SAA, Mst.UH, SZA, PB; Validation and software: SZA, Mst.UH, MS; Correspondence: SAA.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests with the others at all.
Ethical approval
The total research work was conducted under the Ethical Guidelines and Monitoring of Jashore Medical College (JMC), Bangladesh Medical and Dental Council (BMDC) in collaboration with the RPG Authority (Govt. Registration ID: 05-060-06021) under the Project Category H2 (#Project EA No- 19/2022+2023), which was arranged and unconditionally supported by Dr. Sharmin Ahmed (Senior Faculty of Pharmacology Department, Jashore Medical College, Bangladesh). Participants gave written informed consent before data collection began.
Consent for publication
The authors have clear consent for publishing this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jabin, A., Uddin, M.F., Al Azad, S. et al. Target-specificity of different amyrin subunits in impeding HCV influx mechanism inside the human cells considering the quantum tunnel profiles and molecular strings of the CD81 receptor: a combined in silico and in vivo study. In Silico Pharmacol. 11, 8 (2023). https://doi.org/10.1007/s40203-023-00144-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-023-00144-6