Abstract
Chemometrics study that relates biological activity to physicochemical descriptors of a molecule and the prediction of absorption, distribution, metabolism, excretion and toxicity (ADMET) properties in advance are important steps in drugs discovery. In this study, a chemometrics approach was employed on some molecules (inhibitors) of norepinephrine transporter to assess their inhibitory potencies, interactions with the receptor and predict their ADMET/pharmacokinetic properties for identification of novel antipsychotic drugs. The molecules were optimized by using density functional theory at the basis set of B3LYP/6-31G*. The genetic function algorithm technique was used to generate a statistically significant model with a good correlation coefficient R2Train = 0.952 Cross-validated coefficient Q2cv = 0.870, and adjusted squared correlation coefficient R2adj = 0.898. The molecular docking simulation using a neurotransmitter transporter receptor (PDB Code 2A65) revealed that three inhibitors (molecule No 38, 44 and 12) exhibited the highest binding affinity of − 10.3, − 9.9 and − 9.3 kcal/mol, respectively, were observed to inhibit the target by forming strong hydrogen bonds with hydrophobic interactions. The physicochemical and ADMET/pharmacokinetic properties result showed that these three molecules are orally bioavailable, high gastrointestinal absorption, good permeability and non-inhibitors of CYP3A4 and CYP2D6 except for molecule No 38. Also, Molecules No 38 and 44 proved to be non-substrate of P-glycoprotein and nontoxicity to a human ether-a-go-go-related gene with predicted hERG toxicity endpoints (pIC50 < 6) and low ADMET_Risk (< 7.0). The results of this study would provide physicochemical and pharmacokinetics properties needed to identify potent antipsychotic drugs and other relevant information in drug discovery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Psychotic disorders are a group of serious illnesses that affect the mind, and a worldwide mental health challenge which occurred in a person whose mental functioning is impaired to interfere with their capacity to meet the basic demand of life [1]. Psychotic disorders are common to all countries and cause immense human suffering, social exclusion, disability, poor quality of life, staggering economic and social costs. It is estimated that one in every four people has a mental disorder [2]. The combined costs of mental disorder, including loss of productivity, loss of earnings due to illness and social costs, are estimated to total at least USD 113 billion annually [3]. The major depressive disorders (MDDs) had been estimated as the second-largest global burden among all diseases by 2030 which makes the discovery of novel and efficacious antipsychotic drugs very urgent [4]. The symptoms of psychotic disorder include difficulty in concentrating, depressed mood, disrupted sleep pattern, anxiety, suspiciousness, withdrawer from family and friends, delusions, hallucinations, switching topics erratically and suicidal thoughts or action [5].
Norepinephrine (NE) is a neurotransmitter, a crucial neurochemical messenger employed in central noradrenergic and peripheral sympathetic synapses [6] responsible for the reuptake of released norepinephrine (NE) into nerve terminals in the brain. Dysregulation of this neurotransmitter is associated with many debilitating psychotic disorders and mental illnesses [7]. Inhibition of the norepinephrine transporter by NET inhibitors has emerged as important drug targets with a multitude of therapeutic potentials for the treatment of psychiatric disorders and mental diseases [8].
Insilco methods including quantitative structure–activity relationship studies (QSARs), molecular docking and absorption–distribution–metabolism–excretion–toxicity (ADMET) properties/pharmacokinetics studies are key steps employing in drug development and discovery processes [9]. Quantitative structure–activity relationship (QSAR) study finds correlations between biological activities and molecular descriptors of different classes of compound [10], molecular docking simulation elucidates the interactions between the binding molecules, and ADMET/pharmacokinetics assessments provide adequate information on properties that influence absorption, distribution, metabolism, excretion and toxicity of a molecule [9]. It is worthy to note that computer-assisted drug design using Insilco methodology has been of great importance in the identification and development of novel medications for the treatment of antipsychotic diseases and other ailments [11].
The aim of this study is to develop statistically acceptable quantitative structure activity relationship models, using molecular docking simulation to elucidate binding interactions between the molecules and norepinephrine transporter (NET), biological target (receptor) and also to predict their adsorption–distribution–metabolism–excretion–toxicity (ADMET)/pharmacokinetics properties and subsequently compare the results obtained with that of known antipsychotic drugs.
Materials and methods
Dataset collection and geometry optimization
A dataset comprised of 50 molecules of norepinephrine transporter (NET) inhibitors was collected from CHEMBL Database (www.ebi.ac.uk/chembl). The 2D structures of the molecules were generated by using ChemDraw ultra-version 12.0. The 2D structures were subsequently imported into Spartan “14” version 1.4 for geometric optimizations [12] using the density functional theory (DFT) technique of B3LYP at 6-31G* basis set [13] to generate quantum chemical and molecular descriptors.
Division of dataset
The dataset of the studied molecules was partitioned into a training set used to develop the QSAR model and a test set employed for the external validation of the developed model [14] by using the Kennard stone algorithm approach [15] with “Dataset Division GUI 1.2” software.
Model building
A statistical analysis by genetic function approximation (GFA) techniques of the Material Studio software 8.0 version was used to build the QSAR models. GFA has a distinctive attribute to select the basic function genetically generate better models than those made using stepwise regression techniques. The range of variations in this population gives better information on the quality of fit and importance of the descriptors [16]. Friedman’s lack of fit (LOF) was employed to evaluate the quality of the model as a method that measures the fitness of a model. LOF is estimated by this mathematical expression;
where c is the number of basic functions, d is the smoothing parameter, M is the number of samples in the training set, SSE is the sum of square error and p is the sum number of descriptors contained in the model.
Molecular descriptors calculation
Molecular descriptors (OD, ID, 2D and 3D descriptors) are arithmetical values that describe properties of molecules were calculated using paDel-Descriptor software 2.20 version [17] in addition to quantum chemical descriptors generated from the Spartan 14 software.
Assessing quality assurance of the model
Statistical parameters of the model were reviewed and evaluated to ascertain its fitting ability, reliability, predictive ability, stability and robustness of the model generated [18]. The quality assurance of a developed model is guaranteed if the results agreed with global QSAR standard, i.e., R2 > 0.6, R2pred > 0.5, Q2 > 0.6, P (95%) < 0.05, high value of F-test, low values of R2random and Q2random [14].
Validation of the model
Leave-one-out cross-validation technique was employed to determine the predictive power of the model. This was evaluated by using this mathematical expression;
where Ypred, Yexp and \(\overline{Y}_{{{\text{training}}}}\) symbolized the experimental, the predicted and mean values of experimental activity of training set compounds.
Also, the square of the correlation coefficient for the test set (R2test) was evaluated for the predictive capacity of the developed model as part of the external validation technique. The closer the value of R2test value to 1.0, the better the model. The R2test is evaluated by using this mathematical equation;
where Ypred and Ytest are the predicted and experimental activity values of the test set compounds. \(\overline{Y}_{{{\text{training}}}}\) is the mean (average) activity value of the training set.
Y-randomization test
Y-randomization is an important external validation technique to ascertain that a developed QSAR model is strong and reliable and is not inferred by luck [19]. Y-randomization test is performed on the training dataset. The low values of R2 and Q2 is an indication that the model is very robust and highly reliable, and the CR2P value of the model must be greater than 0.5 to pass the Y-randomization test. The CR2P value is calculated by using this mathematical formula;
where CR2P = coefficient of determination for Y-Randomization, R coefficient of correlation for Y-Randomization, Rr Average “R” of random models.
Degree of contribution of selected descriptors
The level of contribution of each descriptor in the model is determined by calculating its standardized regression coefficients bj using this mathematical equation;
bj is the regression coefficient of descriptor j. Sj and Sy are the standard deviations for each descriptor and activity, respectively.
The descriptor of higher absolute standardized coefficient implies greater importance to the rest of molecular descriptors.
Multi-colinearity evaluation
Multi-colinearity estimation among descriptors selected by GFA analysis is evaluated using the variance inflation factor (VIF) by the mathematical expression below;
where R2ij is the correlation coefficient of the multiple regression between the descriptor i and the rest j descriptors in the developed model [20].
Assessment of the applicability domain of the model
Evaluation of the applicability domain of a model is a significant step to confirm that the developed model is capable to make a reliable prediction within the chemical space for which it was developed [21]. To describe the applicability domain of the QSAR model, the leverage approach was employed.
Leverage of a given dataset hi is defined by this mathematical expression;
where \(x_{i}\) the descriptor row is a vector of the considered compound i and hi is the n × k descriptor matrix of the training set compound used to generate the model.
The warning leverage (h*) is the limit of normal values of × outliers and is expressed mathematically as
where n is the number of training compounds and p is the number of predictor variables (descriptors) in the model.
If the leverages hi < h* for the test compounds, it is considered being reliably predicted by the developed model.
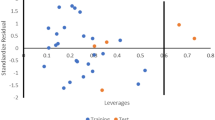
The relevance area of the model in terms of chemical space is visualized by the plot of standardized residuals against leverage values (Williams plot).
Molecular docking simulation
The molecular docking simulation to elucidate the binding interactions between the ligands and the receptor was investigated using the crystal structure of LEUTAA (PDB Code: 2A65), a bacterial homolog of Na+/Cl–-dependent neurotransmitter transporter [22].
Making of ligand and target
All the molecules were optimized and appropriately saved as PDB files. Subsequently, the crystal structure of LEUTAA, a bacterial homolog of Na+/Cl–-dependent neurotransmitter transporter (receptor), was downloaded from the Protein Data Bank Web site (PDB codes 2A65).
Figure 1 displays the prepared structure of the receptor.
The docking of the prepared ligands with the receptor was carried out using AutoDock Vina version 4.0 of Pyrex software, while Discovery Studio software was used to visualize the molecular interactions of the stable complex [23].
ADMET properties and pharmacokinetic studies
The absorption, distribution, metabolism, excretion and toxicity properties predictions, as well as pharmacokinetic evaluation, are major steps for drugs targeting the central nervous system (CNS) because the ability of CNS drugs to penetrate the blood–brain barrier is very critical in metabolism the drug. The ADMET properties predictions and pharmacokinetics study in advance are important methods to save the cost of drug discovery tremendously and also to provide “reality checks” and complimentary second opinions for high throughput assays [24].
In this study, Simulation ADMET Predictor™ 9.0 software, MedChem Designer™ software 5.0 version and SwissADME Web tools were utilized to evaluate ADMET/pharmacokinetics properties, druglikeness and medicinal chemistry friendliness with proficient methods such as the BOILED-Egg, iLOGP and bioavailability radar [25].
Results and discussion
QSAR study was explored to investigate the structure–activity relationship of 50 compounds with distinguishing organic fragments acting as norepinephrine transporter (NET) inhibitors. The nature of models in a QSAR study is expressed by its fitting the data points through regression and making predictions of an isolated dataset.
QSAR on pKi of norepinephrine transporter (NET) inhibitors
A dataset of 50 compounds was divided into a training set of 36 compounds used in developing the model and a test set of 14 compounds was used to evaluate the predictive ability of the QSAR model for the inhibition of norepinephrine transporter. The predicted and experimental activities alongside their residual values are presented in Supplementary Table S1. The low residual values resulted from the experimental and predicted activities is an evidence that the developed model has good predictability.
The genetic algorithm–multiple linear regression (GA-MLR) evaluation prompted the choice of six descriptors, which were eventually used to amassed a linear regression model for calculating pKi of norepinephrine transporter inhibitors within the chemical space of the model. The model with the best statistical significance was selected and represented by Eq. (10):
N is the total number of the datasets, R2 is the squared correlation coefficient and Q2LOO is the squared cross-validation coefficients for leave one out. In the model, the number of ratio of training set data to the ratio number of descriptors present in the model was 6 and in agreement with the Topliss ratio [26]. This implies that the developed model obeyed the QSAR semiempirical rule of thumb [27].The name and the symbol of the descriptors, the standardized regression coefficients (degree of contribution) and percentage contribution of the descriptors are reported in Table 4. The combined presence of 2D and 3D descriptors in the developed model symbolized that these types of descriptors are able to characterize the good antipsychotic properties of the molecules. The sign, magnitude and percentage contribution of each descriptor are not only to give very important information on the direction of influence of the descriptor but also pinpoint the strength of contribution to the activity of the molecule.
The results of the internal and external validations of the model were compiled to Occam's razor rule [28]. Also, generally acceptable QSAR model validation criteria and the validated parameters of the model are presented in Table 1. The values of validation parameters of the model were in excellent agreement with the generally acceptable QSAR model as reported in Table 1. This confirmed the reliability, stability and robustness of the developed model. Pearson’s correlation matrix and other statistical tools employed for validation of the model are reported in Table 2. The low value in correlation coefficients between each pair of descriptors (< 7.0) is a clear indication that there was no pronounced multi-collinearity among the descriptors in the developed model. The variance inflation factor (VIF) values reported in Table 2 were less than 10, and the t-statistics values were greater than 2 for all the descriptors. These are an excellent indications that the developed model was statistically significant, and the descriptors contributed appreciably to the model at a 95% level [29] and they were orthogonal.
The model generated was used to predict the test set data, and the results are reported in Supplementary Table S1. The predicted pKi values for the training and test sets were plotted against the experimental pKi values as shown in Fig. 2. Similarly, the plot of the standardized residuals values for both the training and test sets against the leverage values of the descriptors in the model is shown in Fig. 3. It is clearly noticed from Supplementary Table S1, Fig. 2 and Fig. 3 that the calculated values for the pKi were in good agreement with those of the test set, as a result of this, no any form of error was observed in the model.
QSAR model validation
The internal coherence of the training set was established by using leave-one-out cross-validation technique to ascertain the strength and reliability of the developed model because the candid significance of a QSAR model is not merely their ability to mimic known activities of molecules set by their fitting power (R2), but above all is also their prospective for guessing biological activity accurately. The great value of Q2LOO for pKi of NET inhibitors used (0.8755) proved well of a fully clad internal validation of the model.
The plot of experimental pKi values against predicted pKi values for the training set is presented in Fig. 4. The displayed linear relationship was observed in the plot between the experimental and predicted activities of the training set (R2 = 0.911). The fact that all these results were in agreement with QSAR validation criteria as presented in Table 1 is confirmatory evidence of the reliability, robustness and stability of the developed model [29].
Figure 3 depicts the Williams plot of the NET dataset, in which the standardized residuals for each compound in the dataset were plotted against their leverage values, coming about to acknowledgment of likely outliers and outstanding molecules in the models.
The applicability domain is set up inside a defined domain where all the data points were within the boundary ± 3 for residuals and a leverage threshold h* (\(h* = 3p^{o} /n\) where \(p^{o}\) is the number of model parameters and n is the number of compounds) [19]. From the result, everyone of the molecules of the training set and test set for the dataset was observed to reside inside the domain (square area) and no statistical value far from other compounds (outlier) with standardized residuals > 3 d for the dataset exist.
The percentage of contribution of each descriptor was calculated to determine the relative importance and the contribution of every descriptor in the model. The degree of contribution of each descriptor and variance inflation factor (VIF) of the descriptor were estimated to evaluate the percentage and the significance of the contribution of the descriptors as reported in Tables 2 and 4, respectively. The descriptor GGI10 showed the highest contribution value (27.6%) in the model with VIF value of 4.502 as reported in the two Tables, but its contribution negatively affects the model as observed in Eq. (10) with negative regression coefficient.
The robustness and reliability of the model were evaluated using Y-randomization test to ascertain whether the developed model is by chance correlation or not. The results of this test as reported in Table 3 were also in agreement with QSAR validation criteria reported in Table 1. This is proved that the developed model is robust, good and dynamic. The fact that cR2p value > 0.5 reaffirms that the model possesses good quality assurance and that the model is not only inferred by chance but also very powerful.
Elucidation of descriptors in NET pKi model
By interpreting the molecular descriptors presented in the model (Table 4), it is possible to increase supportive chemical functional groups, fingerprints and pharmacophores into the activities of the NET inhibitors. Therefore, a sufficient interpretation of the QSAR results is reported as follows;
ALogP is a 2D-type molecular descriptor and the first in our QSAR model. It defined as Ghose–Crippen LogKow or Ghose–Crippen–Viswanathan octanol–water partition coefficient. (ALogP) is calculated from the AlogP model consisting of a regression equation based on the hydrophobicity contribution of 115 atom types [30, 31]. AlogP estimates are provided only for compounds having atoms of types C, H, O, N, S, Se, P, B, Si and halogens.
Each atom in every structure is classified into one of the 115 atom types. Then, estimated logP for any compound is given by:
where n is the number of an atom of type i and ai is the corresponding hydrophobicity constant. The list of the atom types with the corresponding hydrophobicity contributions is given under the list of atom-centered fragments. This descriptor tells us the higher the number of heteroatoms in a molecule, the higher the tendency for this molecule to be less hydrophobic. Since the percentage contribution of the descriptor in this model is 13%, it indicates that more than 10% of the bioactivity of a lead molecule will improve the number of heteroatoms present be increased.
AATS7i and ATSC3p are defined as average Broto–Moreau autocorrelation—lag 7/weighted by first ionization potential, and centered Broto–Moreau autocorrelation—lag 3/weighted by polarizabilities, respectively. They are both 2D autocorrelation descriptors and their respective percentage contribution to the models are given as 13 and 16.4%, respectively, in Table 2. The ATS descriptor describes how a property is distributed along with the topological structure. It is a spatial autocorrelation on a molecular graph, which can be used to improve the activity of the compounds by altering the ionization potential and polarizability of the compounds. Since these molecular descriptors contributed positively to the model, the pKi values of the compounds can be improved by adding fragments to the compounds that can increase the polarity of the compounds, thereby creating the charge stability of the ligands’ interaction with the binding sites. GGI10 is a topological charge descriptor defined as the topological charge index of order 10. GGI10 gave the highest contribution in the model, but since its contribution negatively affects the model, the steady reduction in this descriptor value can improve the Ki values of the dataset. The ability of topological charge indices to describe molecular charge distribution has been established by correlating them with the dipole moment of a heterogeneous set of hydrocarbons, so reducing the number of heterogeneous hydrocarbons presently correlated with the dipole moment of the molecule will lead to an increase in the bioactivity of the compounds.
IC2 is defined in Table 2 as the information content index (neighborhood symmetry of two order), and it is a 2D-type information content descriptor. It gave the least contribution to the model, but 10% contribution can be significant depending on the nature of the molecule. The IC2 molecular descriptor suggests that by introducing other bonds at that carbon, the structural complexity of the molecules will be increased and the Shannon entropy will also be increased, thereby easily activating the interactions of the molecule with the binding site.
RDF75u is an RDF descriptor (Radial Distribution Function descriptors), and this descriptor is based on the distance distribution in the geometrical representation of a molecule and constitutes a radial distribution function code (RDF code) that shows certain characteristics in common with the 3D-MORSE code. The radial distribution function in this form meets all the requirements for a 3D descriptor, and it also provides further valuable information such as bond distances, ring types, planar and non-planar systems. This fact is the most valuable consideration for a computer-assisted code elucidation [32]. The positive regression coefficient of this descriptor in the model as contained Eq. (7) with the highest value of the degree of contribution as reported in Table 4 is a good indication of its influential contribution to the antipsychotic property with variation in the bond distance and ring types of the studied compounds.
Docking result
The docking result of this study is presented in terms of binding affinity (kcal/mol) as reported in Supplementary Table S1. All the ligands were docked into the active site of the receptor, using the crystal structure of LEUTAA (PDB: 2A65), a bacterial homolog of Na+/Cl–-dependent neurotransmitter transporter to elucidate binding interactions and mode of inhibition of the target [22].
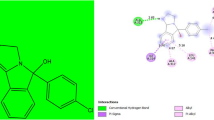
The binding affinity of all the docked molecules ranges from − 5.0 to − 10.3 kcal/mol is reported in Supplementary Table S1. The molecule numbers 38, 44 and 12 were observed to have binding affinity of − 10.3, − 9.9 and 9.3 kcal/mol, respectively, and when visualized and analyzed through the Discovery Studio Visualizer they found to be strongly bonded to the receptor by forming hydrogen bond and also displayed hydrophobic and electrostatic interactions with the target except molecule 38 which could not form hydrogen bond with the receptor as reported in Table 5. The 2D and 3D binding interaction of molecule 12 and 44 is shown in Fig. 5. The amino acid residues in which molecule 12 and 44 formed hydrogen bonds are GLN34 and LYS398 with a bond length of 2.15 Å and 2.63 Å, respectively. The non-hydrogen bond formation displayed by molecule No 38 may be due to its low activity (pKi = 5.084) compared to molecules 12 and 44 with higher activity pKi = 7.383 and 44, pKi = 5.607, respectively. This may suggest that the activity of a molecule could influence the type of bonding interaction that would be observed with a biological target (receptor). However, the highest binding energy displayed by molecule 38 may be due to its large number of hydrophobic interactions and the electrostatic effect due to the presence of fluorine atom, Pi-Cation, Pi-Sigma, Pi-Pi-stacked, Pi-Pi-T-shaped and Pi-Alkyl with amino acid residues (ILE491, ILE410, TRP406, TRP99, PHE494, ARG487, LEU464, ALA464, ILE472) as reported in Table 5.
Physicochemical evaluation of three molecules with highest binding affinity
Physicochemical property is an important attribute of a molecule which influences efficacy, safety or metabolism and can be predicted by using Lipinski’s rule of five (RO5); molecular mass < 500; hydrogen-bond donors (HBD) < 5; hydrogen-bond acceptors (HBA) < 10; and LogP < 5 [33]. The rule suggests that molecules, whose properties fell outside some of these boundaries, would be unlikely to become an orally bioavailable drug [34]. Other physicochemical properties were calculated using Simulation Plus ADMET Predictor™ 9.0. Only the molecule No 38 with the highest binding energy (− 10.3) and lowest molecular weight (329.852 g/mol) among the three selected molecules is fully obeyed the RO5 while the molecule No 12 and 44 have S + LogP values of 5.928 and 5.383 as reported in Table 6. Also, these molecules have a polar surface area less than 70 Å2 and the number of rotatable bonds (RTB) less than 10 except for molecule No 12 that has RTB of 12. These results informed that molecule No 38 and 44 would be orally bioavailable and with good permeability as potential antipsychotic agents [35]. More so, in addition to Log P as a parameter to measure lipophilicity of a substance, Log D which determines the effective lipophilicity a substance was estimated to determine the distribution of a molecule within the body [36]. A Log D value of 1 to 3 and a Log P value greater than 2 are required for a drug-like molecule to cross the blood–brain barrier (BBB) including CNS drugs [37]. These molecules have Log D values between 1–3 except for molecule No 12 that has Log D value slightly above 3 (i.e., 3.236) as reported in Table 6. This result suggests that these molecules possessed a good distribution profile, a property that influences transport processes, intestinal absorption and membrane permeability [36]. All the results obtained for the physicochemical properties of the selected molecules were compared with that of methylphenidate HCl, an approved drug of the antipsychotic disorder as reported in Table 6.
ADMET/pharmacokinetic properties of the selected three molecules
The ADMET/pharmacokinetic properties assessments were carried out using combined Simulation Plus ADMET Predictor™ 9.0 [33] and Swiss ADME tool with a proficient method of Brain Or IntestinaL EstimateD permeation method (BOILED-Egg) [38]. The obtained results for ADMET/pharmacokinetic properties presented in Table 7 revealed that all the selected molecules possessed the attribute for central nervous system (CNS) blood–brain barrier (BBB) penetration and with high gastrointestinal (GI) absorption in comparison with their Log BB values. The BOILED-Egg depicted in Fig. 6 demonstrates the human intestinal absorption (HIA) and the brain access or penetration of a drug molecule. The white region and the yellow region (yolk) are equivalent to the area of the highest probability of being absorbed by the human gastrointestinal tract and highest probability to the brain access (penetration), respectively. It was observed that all three molecules and including methylphenidate HCl (an approved antipsychotic drug) lie within the yellow region (yolk). This is a clear indication that these molecules have a high probability to be absorbed by the gastrointestinal tract and permeate into the brain. More so, the PGP+ (substrates) and PGP− (non-substrates) are symbolized by blue and red dots for molecules assumed to be efflux or non-efflux from CNS by P−glycoprotein, respectively. Molecule No 38 and 44 are PGP−, because of this, they are non-substrates while molecule 12 and that of approved drug (Methylphenidate HCl) are PGP+, and hence, they are substrates of P−glycoprotein.
The effect of cytochrome P450 metabolism for CYP3A4 and CYP2D6 being the most important forms in humans [39] was investigated using both Simulation Plus ADMET Predictor™ 9.0 and Swiss ADME tool. These molecules including methylphenidate HCl were found to be non-inhibitors of CYP3A4 and CYP2D6 except for molecule No 38 that proved to the inhibitor of the enzymes. The ADMET risk and toxicity properties were predicted using Simulation Plus ADMET Predictor™ 9.0 as reported in Table 7. These molecules have ADMET_Risk values less than 7.0 except for molecule No 12. It has been reported that the best potential drug candidates have ADMET_Risk values less than 7.0 [40]. The cardiovascular toxicity was predicted using ADMET Predictor™ 9.0, and the obtained results are reported in Table 7. Certainly, a random block of cardiac human ether-a-go-go-related gene (hERG) channels by a variety of molecules is a major therapeutic challenge with profound impacts on human health [41]. Hence, molecules with hERG toxicity endpoints (pIC50 ≥ 6) are likely to exhibit some hERG toxicity [33]. Luckily enough, all the three molecules and including the approved drug (Methylphenidate HCl) have predicted toxicity endpoints (pIC50 ≥ 6) except for molecule No 12 that has toxicity endpoints (pIC50 = 6.866).
The effect of cytochrome P450 metabolism for CYP3A4 and CYP2D6 being the most important forms in humans [39] was investigated using both Simulation Plus ADMET Predictor™ 9.0 and Swiss ADME tool. These molecules including methylphenidate HCl were found to be non-inhibitors of CYP3A4 and CYP2D6 except for molecule No 38 that proved to the inhibitor of the enzymes. The ADMET risk and toxicity properties were predicted using Simulation Plus ADMET Predictor™ 9.0 as reported in Table 7. These molecules have ADMET_Risk values less than 7.0 except for molecule No 12. It has been reported that the best potential drug candidates have ADMET_Risk values less than 7.0 [40]. The cardiovascular toxicity was predicted using ADMET Predictor™ 9.0, and the obtained results are reported in Table 7. Certainly, a random block of cardiac human ether-a-go-go-related gene (hERG) channels by a variety of molecules is a major therapeutic challenge with profound impacts on human health [41]. Hence, molecules with hERG toxicity endpoints (pIC50 ≥ 6) are likely to exhibit some hERG toxicity [33]. Luckily enough, all the three molecules and including the approved drug (Methylphenidate HCl) have predicted toxicity endpoints (pIC50 ≥ 6) except for molecule No 12 that has toxicity endpoints (pIC50 = 6.866).
Conclusions
This study addressed quantitative structure–activity relationship (QSAR), elucidative binding interactions between the molecules of norepinephrine transporter (NET) inhibitors and the receptor as well as the predictive adsorption–distribution–metabolism–excretion–toxicity (ADMET)/pharmacokinetics properties of the inhibitors. The result of the QSAR modeling was reliable and robust and also satisfied the OECD criteria for model development. The results of internal validation (Q2cv = 0.870) and that of external validation (R2Pred 0.58) suggested the good predictive ability of the model. The results obtained for molecular docking simulation revealed that molecules 38, 44 and 12 have the highest binding affinity of − 10.3, − 9.9 and − 9.3 kcal/mol, respectively, and were found to be firmly bonded to the receptor with the formation of strong hydrogen bonds and hydrophobic interactions of the amino acids of the targets. The physicochemical properties evaluations revealed that the selected three molecules with highest binding energy are orally bioavailable and have good permeability attributes while ADMET properties/pharmacokinetics assessments suggested that these molecules have high chance to be absorbed by the gastrointestinal tract and permeate into the brain as potential antipsychotic agents evidenced from BOILED-Egg diagram. It is hoped these recognized inhibitors would be employed to develop and design novel antipsychotic agents with improved potencies.
References
N. Dessalew, QSAR study on dual SET and NET reuptake inhibitors: an insight into the structural requirement for antidepressant activity. J. Enzym. Inhib. Med. Chem. 24(1), 262–271 (2009)
J. Zhou, NIH Public Access 29(12), 1235–1244 (2006)
D.P. Rice, L.S. Miller, Health economics and cost implications of anxiety and other mental disorders in the United States. Br. J. Psychiatry 173(S34), 4–9 (1998)
W. Xue et al., What contributes to serotonin-norepinephrine reuptake inhibitors’ dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem. Neurosci. 9(5), 1128–1140 (2018)
L.N. Yatham et al., Canadian network for mood and anxiety treatments (CANMAT) and international society for bipolar disorders (ISBD) collaborative update of CANMAT guidelines for the management of patients with bipolar disorder: update 2009. Bipolar Disord. 11(3), 225–255 (2009)
S. Deorah, C.F. Lynch, Z.A. Sibenaller, T.C. Ryken, Trends in brain cancer incidence and survival in the United States: surveillance, epidemiology, and end results program, 1973 to 2001. Neurosurg. Focus 20(4), E1 (2006)
A. Penmatsa, K.H. Wang, E. Gouaux, X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 503(7474), 85–90 (2013)
J. Grouleff, L.K. Ladefoged, H. Koldsø, B. Schiøtt, Monoamine transporters: insights from molecular dynamics simulations. Front. Pharmacol. 6, 235 (2015)
Z. Ye, Y. Yang, X. Li, D. Cao, D. Ouyang, An integrated transfer learning and multitask learning approach for pharmacokinetic parameter prediction. Mol. Pharm. 16(2), 533–541 (2018)
A.-S. Om, J.-C. Ryu, J.-H. Kim, Quantitative structure-activity relationships for radical scavenging activities of flavonoid compounds by GA-MLR technique. Mol. Cell. Toxicol. 4(2), 170–176 (2008)
G.D.J. Davis, A.H.R. Vasanthi, QSAR based docking studies of marine algal anticancer compounds as inhibitors of protein kinase B (PKBβ). Eur. J. Pharm. Sci. 76, 110–118 (2015)
W.J. Hehre, W.W. Huang, Chemistry with computation: an introduction to SPARTAN (Wavefunction, Inc., Irvine, 1995)
R. Bauernschmitt, R. Ahlrichs, Treatment of electronic excitations within the adiabatic approximation of time dependent density functional theory. Chem. Phys. Lett. 256(4–5), 454–464 (1996)
A. Golbraikh, A. Tropsha, Beware of q2! J. Mol. Graph. Model. 20(4), 269–276 (2002)
R.W. Kennard, L.A. Stone, Computer aided design of experiments. Technometrics 11(1), 137–148 (1969)
H. Zeng et al., Cancer survival in China, 2003–2005: a population-based study. Int. J. Cancer 136(8), 1921–1930 (2015)
C.W. Yap, PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32(7), 1466–1474 (2011)
R. Veerasamy, H. Rajak, A. Jain, S. Sivadasan, C.P. Varghese, R.K. Agrawal, Validation of QSAR models-strategies and importance. Int. J. Drug Des. Discov. 3, 511–519 (2011)
K. Roy, S. Kar, P. Ambure, On a simple approach for determining applicability domain of QSAR models. Chemom. Intell. Lab. Syst. 145, 22–29 (2015)
D.E. Arthur, A. Uzairu, P. Mamza, S.E. Abechi, G. Shallangwa, Qsar modelling of some anticancer PGI50 activity on Hl-60 cell lines. Albanian J. Pharm. Pharm. Sci. 3(1), 4–9 (2016)
A. Tropsha, P. Gramatica, V.K. Gombar, The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb. Sci. 22(1), 69–77 (2003)
J. Andersen, K.B. Ringsted, B. Bang-Andersen, K. Strømgaard, A.S. Kristensen, Binding site residues control inhibitor selectivity in the human norepinephrine transporter but not in the human dopamine transporter. Sci. Rep. 5, 15650 (2015)
T. Katsila, G.A. Spyroulias, G.P. Patrinos, M.-T. Matsoukas, Computational approaches in target identification and drug discovery. Comput. Struct. Biotechnol. J. 14, 177–184 (2016)
C. Liao, M. Sitzmann, A. Pugliese, M.C. Nicklaus, Software and resources for computational medicinal chemistry. Future Med. Chem. 3(8), 1057–1085 (2011)
A. Daina, O. Michielin, V. Zoete, SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717 (2017)
J.G. Topliss, R.P. Edwards, Chance factors in studies of quantitative structure-activity relationships. J. Med. Chem. 22(10), 1238–1244 (1979)
S. Van Damme, P. Bultinck, A new computer program for QSAR-analysis: ARTE-QSAR. J. Comput. Chem. 28(11), 1924–1928 (2007)
C.E. Rasmussen, Z. Ghahramani, Occam’s razor, in Advances in neural information processing systems (2001), p. 294–300
A. Tropsha, Best practices for QSAR model development, validation, and exploitation. Mol. Inform. 29(6–7), 476–488 (2010)
V.N. Viswanadhan, A.K. Ghose, G.R. Revankar, R.K. Robins, Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain. J. Chem. Inf. Comput. Sci. 29(3), 163–172 (1989)
A.K. Ghose, G.M. Crippen, Atomic physicochemical parameters for three-dimensional structure-directed quantitative structure-activity relationships I. Partition coefficients as a measure of hydrophobicity. J. Comput. Chem. 7(4), 565–577 (1986)
J. Caballero, M. Fernández, Artificial neural networks from MATLAB® in medicinal chemistry. Bayesian-regularized genetic neural networks (BRGNN): application to the prediction of the antagonistic activity against human platelet thrombin receptor (PAR-1). Curr. Top. Med. Chem. 8(18), 1580–1605 (2008)
D. Lagorce, D. Douguet, M.A. Miteva, B.O. Villoutreix, Computational analysis of calculated physicochemical and ADMET properties of protein–protein interaction inhibitors. Sci. Rep. 7, 46277 (2017)
M. Tareq Hassan Khan, Predictions of the ADMET properties of candidate drug molecules utilizing different QSAR/QSPR modelling approaches. Curr. Drug Metab. 11(4), 285–295 (2010)
P.C. Trippier, Selecting good ‘drug-like’properties to optimize small molecule blood-brain barrier penetration. Curr. Med. Chem. 23(14), 1392–1407 (2016)
D.J. Triggle, J.B. Taylor, Comprehensive Medicinal Chemistry II, vol. 7 (Elsevier, Amsterdam, 2006)
M.J. Waring, Lipophilicity in drug discovery. Expert Opin. Drug Discov. 5(3), 235–248 (2010)
A. Daina, V. Zoete, A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 11(11), 1117–1121 (2016)
K. Tsaioun, S.A. Kates, ADMET for Medicinal Chemists: A Practical Guide (Wiley, Hoboken, 2011)
S. Gabbert, H.-P. Weikard, Sequential testing of chemicals when costs matter: a value of information approach. Hum. Ecol. Risk Assess. Int. J. 19(4), 1067–1088 (2013)
B.O. Villoutreix, O. Taboureau, Computational investigations of hERG channel blockers: new insights and current predictive models. Adv. Drug Deliv. Rev. 86, 72–82 (2015)
Acknowledgements
We wish to acknowledge members of the theoretical and physical chemistry group, the chemistry department, Ahmadu Bello University Zaria. We sincerely appreciate Dr. David Arthur, Abdulfatai Usman and Abdulaleef Jimoh for their technical support and advice in the course of this study.
Funding
The authors received no direct funding for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors pronounced they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human or animal subjects.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Olasupo, S.B., Uzairu, A., Shallangwa, G. et al. QSAR modeling, molecular docking and ADMET/pharmacokinetic studies: a chemometrics approach to search for novel inhibitors of norepinephrine transporter as potent antipsychotic drugs. J IRAN CHEM SOC 17, 1953–1966 (2020). https://doi.org/10.1007/s13738-020-01902-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-020-01902-5