Abstract
In this research, we prepared a new series of the Cu(II) (1) and Ni(II) (2) metal complexes of a tridentate Schiff base ligand, (E)-2-(5-bromo-2-hydroxybenzylideneamino) phenol (H2L). These complexes were characterized by elemental analysis, FT-IR, UV–Vis, and 1H-NMR spectroscopy. The crystal structures of (1) and (2) were determined by X-ray diffraction studies. The single crystal X-ray diffraction analyses revealed that copper(II) cation is five-coordinated and the coordination polyhedron is a slightly distorted square pyramid. Nickel(II), on the other hand, is four-coordinated, and has a regular, square planar geometry. Further discussed were the electrochemical reduction of these complexes. We also analyzed the nature of the metal–ligand bond in the complexes through NBO and EDA analysis. Besides, vibrational sample magnetometer (VSM) revealed complex (1) was ferromagnetic.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Schiff bases, whose metal complexes are of great interest, have often been employed as chelating ligands in the field of coordination chemistry [1,2,3]. They are able to coordinate metals through imine nitrogen and another group, more often than not, linked to aldehyde or ketone [4]. Schiff base metal complexes also find diverse and fruitful applications in industry and daily life [5,6,7,8,9,10]. There exist myriad reports on the biological activities of Schiff base ligands and their metal complexes, including their use for enzyme modeling, and catalytic activity and their function as antimicrobial, antifungal and antitumor agents [11,12,13,14,15]. It has been demonstrated that biological activities of these compounds are influenced significantly by metal coordination [16, 17]. There is also some evidence suggesting that these drugs interact directly with DNA, blocking the activity of DNA–gyrase repair enzymes [18,19,20]. Schiff bases facilitate the process of inducing substrate chirality, tuning the metal-centered electronic factor, enhancing solubility and performing either homogeneous or heterogeneous catalysts [21]. Furthermore, recent studies have witnessed a great deal of interest in the synthesis and characterization of transition metal complexes containing Schiff bases as ligands due to their applications in pharmaceutical fields.
Since the synthesis of new compounds with new properties are important in inorganic chemistry, we have focused on the synthesis and characterization of new Schiff base ligands and their complexes to check out their applications in the future. Considering the remarkable importance of Schiff base complexes and so as to continue our previous work [22,23,24,25], we found it worthwhile to synthesize and characterize new complexes of Ni(II) and Cu(II) with 1-methyl-imidazole and a tridentate Schiff base ligand, obtained by the condensation of 2-aminophenol with 5-bromo-2- hydroxybenzaldehyde (Scheme 1). In addition, a theoretical study was conducted on the structure and nature of the metal–ligand bond in complexes at the BP86/TZ2P(ZORA)//BP86/def2-SVP level of theory.
Experimental
Materials and methods
All starting materials and solvents were purchased from commercial sources and used without further purification. The elemental analyses of carbon, nitrogen and hydrogen atoms were done through the use of a Perkin-Elmer 2400II CHNS-O elemental analyzer. FT-IR spectra were recorded in the range of 4000–400 cm−1 on a FT-IR SHIMADZU spectrophotometer from KBr pellet. UV–Vis spectra were recorded on a UV-1650 PC SHIMADZU spectrophotometer in acetonitrile solution. Using CDCl3 as the solvent, 1H-NMR measurements were performed on NMR BRUKER 400 MHZ spectrometer, at 25 °C. The Schiff base ligand was prepared according to the literature [26]. Also, magnetic measurements were carried out with a vibrating sampling magnetometer (VSM, Model 7400- LakeShore).
Synthesis of the complexes
Synthesis of [CuII(L)(MeIm)(H2O)] (1)
In a typical experiment, a methanolic solution (10 mL) of 0.5 mmol (0.099 g) Cu(CH3COO)2.H2O was added to 0.5 mmol (0.146 g) methanolic solution of ligand. The mixture color turned green. After that, N-methyl imidazole was added dropwise. The reaction mixture was refluxed for 3 h. The green-colored single crystals suitable for X-ray data collection were obtained by slow evaporation of the solution. Yield: 58%. Anal. Calcd. for C17H16BrCuN3O3 (%): C, 44.99; H, 3.55; N, 9.26. Found (%): C, 44.75; H, 3.38; N, 9.25. FT-IR: νmax (cm−1) (KBr): 1608 (νC=N). UV–Vis in CH3CN: λmax (nm) (ɛ, M−1 cm−1): 249 (26,000), 277 (16,100), 304 (8900), 318 (15,050), 427 (13,280).
Synthesis of [NiII(L)(MeIm)] (2)
This complex was prepared by the same procedure applied to complex (1), with the exception that instead of Cu(CH3COO)2.H2O, 0.5 mmol (0.124 g) of the Ni(CH3COO)2.4H2O was used. The red crystals, suitable for X-ray data collection, were obtained by slow evaporation of the methanol solution after 4 days. Yield: 70%. Anal. Calcd. for C17H14BrN3NiO2 (%): C, 47.38; H, 3.27; N, 9.75. Found (%): C, 47.22; H, 3.18; N, 9.70%. FT-IR: νmax (cm−1) (KBr): 1596 (νC=N). UV–Vis in CH3CN: λmax (nm) (ɛ, M−1 cm−1): 238 (43,500), 275 (22,650), 312 (8500), 330 (19,500), 444 (16,200). 1H-NMR (CDCl3, 400 MHz): 8.04 (s, 1H, Hl), 7.65 (s, 1H, Hk), 7.53 (dd, 1H, J1 = 1.2, J1 = 8.4 Hz, Hj), 7.48 (d, 1H, J = 2.4 Hz, Hi), 7.25 (dd, H, J1 = 2.4, J2 = 9 Hz, Hh), 8.61 (s, 1H, Hg), 7.00 (t, 1H, J1 = 1.2, J2 = 7.4 Hz, Hf), 6.84 (t, 1H, J = 1.6 Hz, He), 6.81 (d, 1H, J = 9.2 Hz, Hd), 6.74 (dd, 1H, J1 = 1.2, J2 = 8.4 Hz, Hc), 6.53 (t, 1H, J1 = 1.2, J2 = 8 Hz, Hb), 3.74 (s, 3H, Ha).
X-ray crystallography analyses
The X-ray single crystal data for complexes 1 and 2 were collected at 100(1) K by the ω-scan technique on an Agilent Technologies Xcalibur four-circle diffractometer with Eos CCD detector, equipped with graphite-monochromatized MoKα radiation source (λ = 0.71073 Å). The data were corrected for Lorentz-polarization as well as for absorption effects [27]. Precise unit-cell parameters were determined by a least-squares fit of 2189 (1) and 1478 (2) reflections of the highest intensity, chosen from the whole experiment. The structures were solved with S FT-IR 92 [28] and refined with the full-matrix least-squares procedure on F2 by SHELXL97 [29]. All non-hydrogen atoms were refined anisotropically. Hydrogen atoms were found from water molecule in 1 in the different Fourier map, all other ones were positioned geometrically and refined with a riding model approximation with their displacement parameters constrained to the parent atom with Uiso (H) = 1.2 (CH, CH2, H2O) or 1.5 (CH3) Ueq (C). The summary of crystal data and refinement for (1) and (2) are given in Table 1.
Electrochemistry
Cyclic voltammograms were recorded employing a SAMA Research Analyzer M-500. Three electrodes are utilized in this system, a glassy carbon working electrode, a platinum disk auxiliary electrode and Ag wire as reference electrode. The glassy carbon working electrode was manually cleaned with 1 µm alumina polish prior to each scan. Tetrabutylammonium hexafluorophosphate (TBAH) was used as the supporting electrolyte. Acetonitrile was dried over CaH2. The solutions were deoxygenated for 5 min via purging with Ar. All electrochemical potentials were calibrated against ferrocene–ferrocenium couple under the same conditions [30].
Computational details (theoretical studies)
The geometries of the compounds were optimized without symmetry constraints at the BP86 [1, 2]/def2-SVP [31] level of theory using the Gaussian 03 [32]. It has been shown that BP86 is a suitable level for the calculation of the bonding situation between the M ← L in complexes such as these [33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50]. Vibrational frequency analyses, done at the same level of theory, indicated that the optimized structures are at the stationary points corresponding to the local minima without any imaginary frequency. The bonding analyses pertaining to energy decomposition, were carried out at BP86/TZ2P(ZORA)//BP86/def2-SVP with C1 symmetry. The basis sets for all elements had triple-f quality augmented by one set of polarization functions (ADF basis set TZ2P(ZORA)) with the program package ADF2013.01. EDA calculations were performed to analyse the nature of bonds between the [Ni(MeIm)]2+ and [Cu(MeIm)(H2O)]2+ fragments and L2− ligand (Schiff base ligand) in the foregoing complexes.
Results and discussions
Synthesis
The new metal complexes were synthesized and characterized by elemental analysis, FT-IR, 1H-NMR, UV–Vis and X-Ray diffraction. All complexes were stable at room temperature in the solid state and soluble in common organic solvents (CH2Cl2, EtOH and CH3CN). The spectroscopic data concerning ligand and metal complexes in the “Experimental” section are in good agreement with the expected values.
Spectroscopic characterizations of the complexes
To clarify the mode of complexation and the effect of the metal ion on the ligand, the FT-IR spectra of the free ligand and their Ni(II) and Cu(II) complexes were compared and assigned on the basis of careful and minute comparison. In the IR spectrum of H2L exhibited a broad band characteristic of the OH group at 3300–3500 cm−1. The disappearance of this band in the IR spectra of complexes is indicative of the fact that the Schiff base ligand is coordinated [26]. The IR spectra of these ligands further showed a strong band at 1627 cm−1 attributable to υ(C = N). This band, in the FT-IR spectra of the complexes, shifted to lower frequencies almost between 10 and 30 cm−1 probably indicating the involvement of nitrogen in coordination to the metal ions [23].
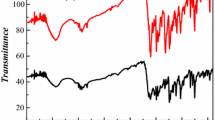
The electronic spectra of Schif base ligand and its complexes were recorded in CH3CN (Fig. 1). As seen, the Schiff base ligand shows a strong band at 362 nm which can be associated to n → π* transition of the azomethine chromophore. This band disappears in complexes after bonding Schiff base ligand to metal center [51]. All the bands in the 200–300 nm region are attributed to the π → π* transitions of the aromatic rings and the azomethine group. The ligand bands shift to longer wavelengths in the metal complexes as compared to their position in the free ligand which indicates the bond between Schiff base and metal center [52]. Complexes (1) and (2) show bands in 410–470 nm region assigned to the CT transition.
As shown in Fig. 2, the 1H-NMR spectral data of the complex was recorded in CDCl3 with the chemical shifts expressed in ppm downfield from tetramethylsilane and presented in “Experimental” section. The 1H-NMR spectra of complex 2 indicate sharp singlet signals (s) at around 8.03 ppm due to the presence of azomethine protons, Hl. Moreover, the singlets at 3.74 ppm are assigned to CH3 protons of N-methyl-imidazole, Ha. The aromatic protons of coordinated Schiff base and the amine ligands, Hb,c,d,e,f,g,h,i,j,k, appear in the appropriate region of 6.51–7.65 ppm in complex 2.
Description of the crystal structures
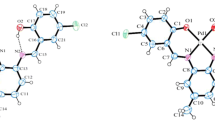
The perspective views of the complexes 1 and 2 are shown in Figs. 3 and 4, respectively. Selected bond lengths and angles for both complexes are listed in Table 2. In 1, Cu(II) center is in a five-coordinate environment. The coordination polyhedron is a distorted square pyramid, with two oxygen and nitrogen atoms from the Schiff base ligand and N atom from N-methyl imidazole creating the base, and oxygen atom from a water molecule at the axial position. The four basal atoms are making quite a good plane (maximum deviation of 0.095(2) Å), and the Cu–O1W direction is almost perfectly perpendicular to this plane (88.5°). The axial Cu1–O1W bond distance of 2.335(2) Å is significantly larger than the basal Cu1–O1 and Cu1–O10 bond distances of 1.969(2) and 1.910(2) Å, respectively. The Cu–N and Cu–O bond distances (Table 3) agree well with similar other square pyramidal Schiff base complexes of copper(II) [53, 54].
The coordination number of Ni(II) ion in 2 is 4 (one nitrogen and two phenolic oxygen atoms from the tridentate ligands, plus one nitrogen atoms from N-methyl imidazole molecules) and the geometry is square-planar. Maximum deviation from the least-squares plane defined by four coordination centers is as small as 0.066(2) Å and the Ni ion lies exactly in this plane (deviation of 0.004(2) Å). Table 3 shows that the angles within coordination square are slightly different—in fact, such a slightly distorted square planar geometry is similar to previously reported analogs [55, 56].
In both complexes, the Schiff base ligand molecule is almost planar, the dihedral angle between the planes of terminal rings is 8.0(3)° in 1 and 7.7(4)º in 2.
In the crystal structure of 1—thanks to the presence of water molecule—O–H(water)···O(ligand) hydrogen bonds connect molecules into infinite chains along y-direction (Table 3; Fig. 5). Structure of 2 lacks strong hydrogen bond donors, so the complex molecules create the layers, only very loosely interacting with one another (Fig. 6).
Electrochemical studies
The cyclic voltammetry of the complexes (1, 2) was conducted at 25 °C under an argon atmosphere using acetonitrile solvent containing 0.1 mol dm−3 TBAH as the supporting electrolyte and complex concentrations of about 4 × 10−3 mol dm−3. In general, redox data are used to identify the model system among the synthetic complexes. In the copper complex, the redox couple Cu(II)/Cu(I) is important and is known to be influenced by such ligand factors as the nature of donor atoms and their structural arrangements around the copper ion [57]. The copper complex (Fig. 7) shows an irreversible redox couple with a cathodic and anodic peak potential at − 1.08 and − 0.93 V, respectively [58]. The cyclic voltammogram of Ni(II) complex (Fig. 8) shows an anodic peak at Epa = 0.82 V corresponding to NiII → NiIII [59,60,61,62].
Theoretical studies
The geometry of the metal complexes of both [Cu(L)(MeIm)(H2O)] (1) and [Ni(L)(MeIm)] (2) complexes was specified by the X-ray crystal structure analysis, and fully optimized at BP86/def2-SVP level of theory. The obtained optimized geometry of both above complexes were similar to those derived by X-ray crystal structures (See Fig. 9). Table 4 shows the selected calculated bond lengths and bond angles of both complexes. With the help of NBO analysis, we further investigated the nature of bonds between two interacting fragments, [Cu(MeIm)(H2O)]2+ and [Ni(MeIm)]2+ and L2− ligand (the studied Schiff base ligand). The Wiberg bond indices (WBIs) for M → L (M = Cu2+,Ni2+) bonds were calculated, which corresponding values are given in Table 4. The values of WBIs for M → N bonds in the complexes are almost similar, yet those of M → O bond lengths obtained for [Ni(L)(MeIm)] complex are greater than the corresponding values of [Cu(L)(MeIm)(H2O)] complex. Also, in the complexes, the value of M → N bonds obtained from Schiff base ligand are slightly higher than that obtained from imidazole ligand.
Further studied were the values of natural charges on M metal ions and N and O atoms in Schiff base ligand (L) and imidazole ligands (see Table 5).
Also, the values of charge transfer from L2− ligand (Schiff base ligand) to [Cu(MeIm)(H2O)]2+ and [Ni(MeIm)]2+ fragments in the latter complexes are about − 0.96 and − 0.93 e, respectively.
Morokuma [63] and Ziegler et al. [64] have recently presented the “energy decomposition analysis” (EDA) method, where a quantitative computational pattern explains the strength of M ← L σ donation and M → L back bonding in the main group and transition metal complexes with different types of ligands [33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50, 65,66,67,68,69].
To better fathom the nature of the bonding situation between [Cu(MeIm)(H2O)]2+ and [Ni(MeIm)]2+ fragments and Schiff base ligand L in the complexes, the energy-decomposition analysis (EDA) was carried out at BP86/TZ2P(ZORA)//BP86/def2-SVP with C1 symmetry. The results indicated that the ∆Eint for [Ni(L)(MeIm)] complex is about 545.8 kcal mol−1 and more than the corresponding value for [Cu(L)(MeIm)H2O] complex (see Table 6). The breakdown of the ΔEint values into the Pauli repulsion, ΔEPauli and the three attractive components shows that roughly 65% come from the electrostatic attraction (ΔEelstat) while ~ 34% come from the orbital term ΔEorb in [Cu(L)(MeIm)(H2O)] and[Ni(L)(MeIm)] complexes. In addition, the values of ∆Eelstat show that the nature of M → N bonds in the complexes is more electrostatic. The covalent bonding between the two interacting fragments, [Cu(MeIm)(H2O)]2+ and L2− in [Cu(L)(MeIm)(H2O)] complex and [Ni(MeIm)]2+ and L2− in [Ni(L)(MeIm)] complex becomes visible by the calculated deformation densities Δρ, which are associated with the significant orbital interactions between the corresponding fragments. Figures 10 and 11 show important deformation densities (Δρ) and related energy values, providing around 61 and 73.5% of the overall orbital interactions for both [Cu(L)(MeIm)(H2O)] and [Ni(L)(MeIm)] complexes, respectively. Figure 10 illustrates the deformation densities Δρ1α and 1β, Δρ2β, Δρ3β, Δρ5α, Δρ6α and 6β, and Δρ7β,which come from M ← L σ donation from the lone pair of N and O atoms of L2− ligand (Schiff base ligand) to Cu metal ion in [Cu(L)(MeIm)(H2O)] complex. In the last figure, the deformation densities, Δρ3β, and Δρ4β represent the M ← L π donation and Δρ4α and Δρ5β represent the π back-donation, respectively (See Fig. 8). On the other hand, the visual inspection of Fig. 11 indicates that Δρ1α and 1β, Δρ4β, Δρ5α, Δρ6α, Δρ7β, Δρ8α and Δρ8β come from the M ← L σ donation from the lone pair of N and O atoms of L2− ligand (Schiff base ligand) to Ni metal ion in [Ni(L)(MeIm)] complex. Furthermore, the deformation densities Δρ3α, Δρ3β, and Δρ5β represent the M ← L π donation and Δρ4β indicates the π back-donation. Note that the colour in Figs. 8 and 9 denotes the charge flow, which is from red to the blue region.
Magnetic measurement
The magnetic characterization of [Cu(L)(MeIm)(H2O)] (1) complex was examined using vibration sampling magnetometer (VSM). The magnetic behavior of the complex in the M–H [M—magnetization (memu/g) and H—magnetic field (Gouss)] curve are shown in Fig. 12. The fine shape of the hysteresis loop is a characteristic of a weak ferromagnetic behavior.
Conclusions
Two new Schiff base complexes were synthesized and characterized by the spectral and analytical techniques. The crystal structures of complexes (1) and (2) were specified by X-ray crystallography. The electrochemical reduction of these complexes at a carbon glass electrode in CH3CN solution indicates that the redox reaction of Ni(III)/Ni(II) and Cu(II)/Cu(I) is irreversible. The bonding situation between the two interacting fragments, [Ni(MeIm)]2+ and L2− in [Ni(L)(MeIm)] complex and [Cu(MeIm)(H2O)]2+ and L2− in [Cu(L)(MeIm)(H2O)] complex was analyzed by NBO and energy-decomposition analysis (EDA), and its natural orbitals for chemical valence variation (EDA–NOCV). The findings indicated that the values of WBIs for M → N bonds in the complexes are almost similar but the values of M → O bond lengths obtained for [Ni(L)(MeIm)] complex are slightly more than the corresponding values for [Cu(L)(MeIm)(H2O)] complex. Ultimately, the ETS–NOCV schemes demonstrated that the ΔEorb term mainly arises from M ← L σ donation, while a smaller contribution comes from M ← L π donation and M → L π back-donation. Magnetization measurements of complex (1) showed a weak ferromagnetic behavior.
References
H.J. Temel, Coord. Chem. 57, 723–729 (2004)
H. Temel, B. Ziyadanogullari, I. Aydin, F. Aydin, J. Coord. Chem. 58, 1177–1185 (2005)
W.K. Dong, Y.X. Sun, Y.P. Zhang, L. Li, X.N. He, X.L. Tang, Inorg. Chim. Acta. 362, 117–124 (2009)
S. Shit, S. Sen, S. Mitra, D.L. Hughes, Transit. Met. Chem. 34, 269–274 (2009)
S. Dutta, P. Biswas, Polyhedron. 31, 110–117 (2012)
H.W.J. Yang, J.F. Maa, J.Y. Lee, T.F. Xie, Polyhedron 31, 136–142 (2012)
R.B. Samulewski, J.D. da Rocha, R. Stieler, E.S. Lang, D.J. Evans, G. Poneti, O.R. Nascimento, R.R. Rbeiro, F.S. Nunes, Polyhedron 30, 1997–2006 (2011)
Q. Cheng, Z. Pan, H. Zhou, J. Chen, Inorg. Chem. Commun. 14, 929–933 (2011)
S. Anbu, M. Kandaswamy, P. Suthakaran, V. Murugan, B.J. Vaeghese, Inorg. Biochem. 103, 401–410 (2009)
V.P. Garcia, D.V. Yazigi, A. Cabrera, P.V. Galvez, M. Arriagada, D.R. Leon, N. Pizarro, A. Zanocco, E. Spodine, Polyhedron 28, 2335–2340 (2009)
T.M. Tallon, Synthesis of imidazole schiff base ligands, their silver(I) complexes and their activities against candida (Albicans Department of Chemistry The National University of Ireland, Mynooth Co, Mynooth, 2012)
M. Hasanzadeh, M. Salehi, M. Kubicki, S.M. Shahcheragh, G. Dutkiewicz, M. Pyziak, A. Khaleghian, Transit. Met. Chem. 39, 623–632 (2014)
N. Charefa, F. Sebtia, L. Arrara, M. Djarmounia, N. Boussoualima, A. Baghiania, S. Khennoufb, A. Ouraric, M.A. Damend, M.S. Mubarakd, D.G. Peters, Polyhedron 85, 450 (2015)
S. Chandraa, S. Agrawal, Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 124, 564 (2014)
M. Tyagia, S. Chandraa, J. Akhtarb, D. Chand, Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 118, 1056 (2014)
H.R. Park, T.H. Kim, K.M. Bark, J. Euro, Med. Chem. 37, 443–460 (2002)
R. Gust, I. Ott, D. Posselt, K. Sommer, J. Med. Chem. 47, 5837 (2004)
A. Tarushi, C.P. Raptopoulou, V. Psycharis, A. Terzis, G. Psomas, D.P. Kessissoglou, Bioorg. Med. Chem. 18, 2678–2685 (2010)
J.H. He, D.R. Xiao, H.Y. Chen, D.Z. Sun, S.W. Yan, X.W.Z. Li, Q.L. Ye, E.B. Wangb, Solid State Chem. 198, 279–288 (2013)
J.L. Zeng, F.R. Zhu, S.B. Yu, F.Z. Zeng, J. Wu, L. Tan, S.H.H. Zheng, N.N. Wang, L. Zhang, Y. Jian, J. Trans. Metal chem. 37, 765–770 (2012)
N. Soltani, M. Behpour, S.M. Ghoreishi, H. Naeimi, Corros. Sci. 52, 1351–1361 (2010)
M. Salehi, M. Amirnasr, S. Meghdadi, K. Mereiter, H.R. Bijanzadeh, A. Khaleghian, Polyhedron 81, 90–97 (2014)
R. Taherlo, M. Salehi, Inorg. Chim. Acta. 418, 180–186 (2014)
M. Salehi, M. Hasanzadeh, Inorg. Chim. Acta. 426, 6–14 (2015)
R. Fekri, M. Salehi, A. Asadi, M. Kubicki, Polyhedron, 128, 175–187 (2017)
E. Labisbal, L. Rodrĺguez, A. Vizoso, M. Alonso, J. Romero, J.-A. Garcĺa-Vázquez, A. Sousa-Pedrares, A. Sousa, Z. Anorg. Allg. Chem. 631, 2107–2114 (2005)
Oxford Diffraction, CRYSALIS PRO, Version 1.171.33.36d (Oxford Diffraction Ltd., Yarn ton, Oxford shire, 2009)
A. Altomare, G. Cascarano, C. Giacovazzo, A. Guagliardi, J. Appl. Crystallogr. 26, 343–350 (1993)
G.M. Sheldrick, Acta Crystallogr. Sect. A64, 112–122 (2008)
N.G. Connelly, W.E. Geiger, Chem. Rev. 96, 877–910 (1996)
A. Schäfer, H. Horn, R. Ahlrichs, J. Chem. Phys. 97, 2571–2578 (1992)
M.J. Frisch et al., Gaussian 03, Revision C.01 (Gaussian, Inc., Wallingford, 2003)
S.J. Sabounchei, M. Pourshahbaz, S. Salehzadeh, M. Bayat, R. Karamia, M. Asadbegy, H.R. Khavasi, Polyhedron 85, 652–664 (2015)
S.J. Sabounchei, H. Nemattalab, S. Salehzadeh, S. Khani, M. Bayat, H.R. Khavasi, Polyhedron 27, 2015–2021 (2008)
H. Keypour a, N. Rahpeyma, M. Rezaeivala b, P. Arzhangi, M. Bayat, L. Valencia, Y. Elerman, O. Büyükgüngör, Polyhedron 51, 117–122 (2013)
H. Keypour a, A. Shooshtari, M. Rezaeivala, M. Bayat, H. Amiri, Rudbari, Inorg. Chim. Acta 440, 139–147 (2016)
H. Keypour a, M. Rezaeivala, A. Ramezani-Aktij, M. Bayat, N. Dilek, H. Ünver, Mol. Struct. 1115, 180–186 (2016)
M. Bayat, M. Hatami, Polyhedron 110, 46–54 (2016)
S.J. Sabounchei, A. Yousefi, M. Ahmadianpoor, A. Hashemi, M. Bayat, A. Sedghi, F.A. Bagherjeri, R.W. Gable, Polyhedron 117, 273–282 (2016)
H. Keypour, M. Liyaghati‑Delshad, M. Rezaeivala, M. Bayat, J. Iran. Chem. Soc. 12, 621–630 (2015)
S.J. Sabouncheia, S. Samiee, M. Pourshahbaza, S. Salehzadeha, M. Bayat, D. Nematollahia, R. Karamian, M. Asadbegy, J. Chem. Res. 38, 35–40 (2014)
S.J. Sabouncheia, M. Zamaniana, M. Pourshahbaza, M. Bayat, R. Karamian, M. Asadbegy, J. Chem. Res. 40, 130–136 (2016)
S.J. Sabounchei, M. Ahmadianpoor, A. Yousefi, A. Hashemi, M. Bayat, A. Sedghi, F. Akhlaghi, R.W. Bagherjeri, Gable, RSC Adv. 6, 28308–28315 (2016)
S. Salehzadeh, M. Khalaj, S. Dehghanpour, M. Bayat, J. Iran. Chem. Soc. 11, 9–16 (2014)
M. Bayat, F. Amraie, S. Salehzadeh, J. Theor.Comp. Chem. 15, 16500321–165003219 (2016)
S.J. Sabounchei, M. Sarlakifa, S. Salehzadeh, M. Bayat, M. Pourshahbaz, H.R. Khavasi, Polyhedron 38, 131–136 (2012)
M. Bayat, M. von Hopffgarten, S. Salehzadeh, G. Frenking, J. Organomet. Chem. 696, 2976–2984 (2011)
M. Bayat, S. Salehzadeh, G. Frenking, J. Organomet. Chem. 697, 74–79 (2012)
M. Bayat, N. Ahmadian, Polyhedron 96, 95–101 (2015)
M. Bayat, N. Ahmadian, J. Iran. Chem. Soc. J Iran Chem Soc 13, 397–402 (2016)
M. Galini, M. Salehi, M. Kubicki, A. Amiri, A. Khaleghian, Inorg. Chim. Acta 461, 167–173 (2017)
M. Jafari, M. Salehi, M. Kubicki, A. Arab, A. Khaleghian, Inorg. Chim. Acta 462, 329–335 (2017)
V. Philip, V. Suni, M.R.P. Kurup, M. Nethaji, Polyhedron 24, 1133–1142 (2005)
M. Pragathi, K.H. Reddy, Inorg. Chim. Acta 413, 174–186 (2014)
M. Mariappan, M. Suenaga, A. Mukhopadhyay, B.G. Maiya, Inorg. Chim. Acta 390, 95–104 (2012)
Y. Huang, T. Liu, J. Lin, J. Lü, Z. Lin, R. Cao. Inorg. Chem. 50, 2191 (2011)
(a) L. Casella, M. Gullotti, A. Pintar, F. Pinciroli, R. Vigano, P. Zanello, J. Chem. Soc. Dalton Trans. 1161 (1989); (b) S. Sevagapandian, G. Rajagopal, K. Nehru, P.R. Athappan, Trans. Met. Chem. 25, 388–393 (2000)
V.T. Kasumov, F. Koksal, A. Sezer, Polyhedron 24 (2005) 1203–1211; (b) P. Patel, P.K. Bhattacharya, J. Inorg. Biochem. 54, 187–197 (1994)
I.C. Santos, M. Vilas-Boas, M.F.M. Piedade, C. Friere, M.T. Duarte, B. De Castro, Polyhydron 19, 655–664 (2000)
L. Gomes, E.S. Periera, B. De Castro, J. Chem. Soc. Dalton Trans. 15, 1373–1379 (2000)
A. Kapturkiewicz, B. Behr, Inorg. Chim. Acta 69, 247–251 (1983)
C. Freire, B. De Castro, J. Chem. Soc. Dalton Trans. 9, 1491–1498 (1998)
T. Ziegler, A. Rauk, Theor. Chim. Acta. 46, 1–10 (1977)
G. Frenking, K. Wichmann, N. Fröhlich, C. Loschen, M. Lein, J. Frunzke, V.C.M. Rayón, Coord. Chem. Rev. 238–239, 55–82 (2003)
M. Lein, A. Szabo, A. Kovacs, G. Frenking, Farad. Dis. 124, 365–378 (2003)
M. Lein, G. Frenking, Chap. 13 (Elsevier, Amsterdam, 2005), pp. 291–372
M. Bayat, S. Kamali, J. Mol. Liq. 222, 953–962 (2016)
M. Bayat, E. Soltani, Polyhedron, 123, 39–46 (2017)
M. Bayat, A. Sedghi, L. Ebrahimkhani, S.J. Sabounchei, Dalton Trans. 46, 207–220 (2017)
Acknowledgements
We thank Semnan University for supporting this study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material. Crystallographic data (excluding structure factors) for the structural analysis have been deposited with the Cambridge Crystallographic Data Centre, Nos. CCDC 1044763 (1) and CCDC 1044764 (2). Copies of this information may be obtained free of charge from: The Director, CCDC, 12 Union Road, Cambridge, CB2 1EZ, UK. Fax: +44(1223)336-033, email:deposit@ccdc.cam.ac.uk, or www: www.ccdc.cam.ac.uk.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Salehi, M., Faghani, F., Kubicki, M. et al. New complexes of Ni(II) and Cu(II) with tridentate ONO Schiff base ligand: synthesis, crystal structures, electrochemical and theoretical investigation. J IRAN CHEM SOC 15, 2229–2240 (2018). https://doi.org/10.1007/s13738-018-1412-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-018-1412-1