Abstract
The clinicians usually desire to know the shape of the liver during treatment planning to minimize the damage to the surrounding healthy tissues and hepatic vessels, thus, building the geometric model of the liver becomes paramount. There have been several liver image segmentation methods to build the model over the years. Considering the advantages of conventional image segmentation methods, this paper reviews them that spans over last 2 decades. The review examines about twenty-five automated and eleven semi-automatic approaches that include Probabilistic atlas, K-means, Model and knowledge-based (such as active appearance model, live wire), Graph cut, Region growing, Active contour-based, Expectation Maximization-based, Level sets, Laplacian network optimization, etc. The main contribution of this paper is to highlight their clinical suitability by providing their advantages and possible limitations. It is nearly impossible to assess the methodologies on a single scale because a common patient database is usually not used, rather, diverse datasets such as MICCAI 2007 Grand Challenge (Sliver), 3DIRCADb, Zhu Jiang Hospital of Southern Medical University (China) and others have been used. As a result, this study depends on the popular metrics such as FPR, FNR, AER, JCS, ASSD, DSC, VOE, and RMSD. offering a sense of efficacy of each approach. It is found that while automatic segmentation methods perform better technically, they are usually less preferred by the clinicians. Since the objective of this paper is to provide a holistic view of all the conventional methods from clinicians’ stand point, we have suggested a conventional framework based on the findings in this paper. We have also included a few research challenges that the readers could find them interesting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Although the term “segmentation” sounds simple, it is important in hepatic disease diagnosis and treatment planning; it can well be used in intra-operative navigation and registration of multimodal images/instruments (Mohanty and Dakua 2022) during the actual procedure. For instance, holding a physical organ phantom in hand for surgical planning is way better than just imagining the organ by looking at its imaging modality, say computed tomography (CT). Therefore, accurate organ segmentation is presently considered indispensable in typical surgical pipelines (Hsu and Chen 2008; Lee et al. 2005; Bezdek et al. 1993; Clarke et al. 1995; Morrison and Attikiouzel 1994; Lim and Pfefferbaum 1989; Harris et al. 1991; Amartur et al. 1992; Ozkan et al. 1993). To be further specific, appropriate resection of a complex liver lesion depends on the degree of accurate visualization of liver anatomy. The experienced and expert clinicians can deal with such cases easily. However, when it comes to novice surgeons or residents, they need proactive supervision. In such cases, a physical 3D phantom is quite helpful. To have a reliable 3D model of the liver, the segmentation of the liver is needed and it needs to be highly accurate. Over the last couple of decades, researchers have been driven to achieve the most efficient segmentation technique that would allow the clinicians to have an easy access to organ measurements and visualize. For this reason, the computer vision research community has put a lot of effort in developing image segmentation methods over the years (Lin et al. 1992; Brandt et al. 1994; Hall et al. 1992; Ortendahl et al. 1985; Kohn et al. 1991; Liang et al. 1992; Liang 1993; Liang et al. 1994; Choi et al. 1991; Taxt and Lundervold 1994; Lundervold and Storvik 1995; Santago and Gage 1993). There have been some surveys as well: Heimann et al. present a review paper that compares and evaluates the liver segmentation papers from CT scans (Heimann et al. 2009). They find the semi-automatic methods better over automatic ones. Campadelli et al. present a detailed review on the automatic methods and propose a new method (Campadelli and Esposito 2009). Another review paper suggests that the segmentation performance varies with change in the input data (Dakua 2013a). Yusuf et al. assess the risks of using computer generated segmentation software (Akhtar et al. 2021) in treatment planning of liver lesions; they find that the lesion relapses in future; that means the segmentation software has to be accurate. Pragati et al. assess the feasibility and efficiency of fusion for post ablation assessment of liver neoplasms (Rai et al. 2021). Anchal et al. present a review on the present therapeutics targeting liver lesions and find that image fusion between two imaging modalities can provide better information about the lesion than a single imaging modality (Dakua and Nayak 2022). Mohammad et al. survey various image segmentation methods and conclude that artificial intelligence-based methods could be effective from clinical standpoint (Ansari et al. 2022b; Singh et al. 2023). Thus, there have been some surveys on segmentation methods, however, in most of the surveys, only a limited number of studies are included, some with the number of methods, some with a section of methods, some with a certain objective: for instance in Heimann et al. (2009), the whole survey was limited to a conference. Campadelli and Esposito (2009) emphasize mostly on the automatic methods that might not be fair from clinicians’ view point, because the clinicians sometimes want to adjust the delineation based on the clinical need and their clinical expertise. The survey (Dakua 2013a) does focus on both semi-automated and automated methods, but again, this has focussed on the performance of a method when the input data is varied. The authors in Akhtar et al. (2021) focus on the possible relapse of a lesion over the time if a computer aided diagnosis (CAD) software is considered during the treatment. Similarly, the other surveys (Dakua and Nayak 2022) and (Rai et al. 2021) focus on the fusion aspect. The present survey paper is bit different than the existing ones; to the best of our knowledge, there has been probably no study in public domain that depicts the clinical utilizations of the conventional methods covering such a wide spectrum of duration (from 2000 to 2022). Despite having several segmentation tools, there is no method that can be applied to all human organs effectively (Al-Kababji et al. 2022), there is not a generic image segmentation tool that can be applied to the multi-modal images of a human organ such as CT, MRI, and US. Thus, in this paper, we have presented the potential utilities of each method with respect to their clinical usability, which no other survey paper has probably reported. The abbreviations used in the paper are given in Table 1.
1.1 Clinical relevance of liver CT segmentation
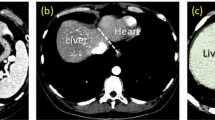
The liver being an important organ of our body, the segmentation techniques are essential for the measurement of liver volume (Nakayama et al. 2006), hepatic surgical planning for hepatocellular carcinoma (as shown in Fig. 1) (Fan et al. 2000), study of anatomical structure, localization of pathology, diagnosis, and computer integrated surgery (Halabi et al. 2020). Thus, accurate segmentation of liver, tumor(s), and arteries from the imaging volumes is among the primary goals of computerized image processing (Al-Kababji et al. 2023).
The primary criteria for a segmentation method should be its simplicity, user friendliness, accuracy, and fast execution; furthermore, it should be compatible to the clinical requirements. Citing the emergence of new diseases, varieties, and the complexities, the medical diagnoses are probably incomplete in the absence of substantial imaging modalities such as positron emission tomography (PET), computed tomography (CT), ultrasound imaging (US), or magnetic resonance imaging (MRI) (Dakua and Sahambi 2009). Each imaging technology has its own merits and demerits, for example, despite its non-intrusive nature and lack of radiation emission, US is precisely operator dependent. MRI, with its noninvasive nature and high tissue contrast, can differentiate and detect tumors present in the liver accurately, yet, the examination is highly expensive. PET successfully creates a three-dimensional image or representation of the body’s functioning operations. Concurrently, one of the biggest drawbacks of PET is that the majority of the probes need to be created using a cyclotron, making it more expensive. CT, on the other hand, with less time and expense provides the details of the organ, in addition to providing finer spatial resolution and advanced signal-to-noise ratio. Therefore, US, PET and MRI are not as popular as CT.
In this paper, the main contributions are as follows:
-
1.
we review the popular liver image segmentation algorithms from CT scans over last 20 years,
-
2.
we present the suitable clinical environments for each segmentation, where the segmentation method can be effective, and
-
3.
finally, we critically discuss each method, the rationale for not being able to provide a robust solution, and a potential solution.
The article comprises of five sections. Section 2 enunciates the inclusion and exclusion criteria to choose the methods in this review. Section 3 describes the methods individually. The quantitative results by the methods and their suitability are detailed in Sect. 4. In Sect. 5, an introspect of the precedence, drawbacks of segmentation techniques, and the outlook of the review are included, while Sect. 6 concludes the paper.
2 Inclusion and exclusion criteria
This section includes the quantitative measures to estimate the quality of segmentation and various existing segmentation methods from CT scans (Dakua 2013a, b, 2014; Zhai et al. 2018; Dakua et al. 2018).
2.1 Journals of interest
The research on liver segmentation has gained momentum after early 90 s (Pham et al. 2000) and Pham et al. report these methods extensively up to the year 2000. After the year 2000 until 2022, there have been several review papers in bits and pieces, but to the best of our knowledge, there is not any review paper that has dissected all the conventional segmentation approaches. All the conventional methods, in a single paper, should probably be able to convey the advantages and disadvantages of these methods to the readers providing a holistic view about these methods. Taking these points into account, we decided to focus on liver segmentation methods from CT scans after the year 2000. Our current research objective is to find the difficulties in deciding a generic method for liver region extraction. In this review, we have included all the segmentation algorithms from high impact factor journals and conferences ensuring the credibility of the findings; the venues include IEEE Transactions on Biomedical Engineering, IEEE Transactions on Medical Imaging, IEEE Transactions on Pattern Analysis and Machine Intelligence, IEEE sensor journals, IEEE Transactions on Image Processing, Computer Vision and Image Understanding, Computerized Medical Imaging and Graphics, Artificial Intelligence Review, Computer in Biology and Medicine, Artificial Intelligence in Medicine, European Journal of Radiology, Academic Radiology, International Journal for Computer Assisted Radio Surgery, Medical Physics, and others because they are believed to have been proved very much influential to the scientific community. The outcomes of this survey are based on the findings from these high-impact venues ensuring its quality, which the researchers could leverage. We have included the studies/papers in this survey by following a process as shown in Fig. 2.
2.2 Standard segmentation evaluation metrics
There are several evaluation metrics in the literature to evaluate the segmentation efficacy (geometric model of a liver is shown in Fig. 3); in this section, we discuss the most preferred ones highlighting the notations, and the significance (Dakua et al. 2018; Dakua and Abi-Nahed 2013; Dakua 2017).
2.2.1 Notations
The notations are:
-
A refers to the ground-truth label voxels set
-
B is the predicted voxels set by the created models
-
\(|\cdot |\) is the set cardinality
-
\(||\cdot ||\) represents the Euclidean distance
-
\(S(\cdot )\) indicates the set of surface voxels
-
True positive (TP) is the set of correctly classified tissue of interest (TOI) pixels/voxels
-
True negative (TN) is the set of truly classified background pixels/voxels
-
False positive (FP) is the set of incorrectly classified background pixels/voxels
-
False negative (FN) is the set of incorrectly classified TOI pixels/voxels
2.2.2 Jaccard index (JI)
JI is a fundamental metric to understand how close is the generated prediction in overlapping with the ground-truth label. It is also known as intersection-over-union (IoU) metric:
Intuitively, perfect prediction is when JI is equal to 1, meaning that \(|A \cap B|\) is the same as \(|A \cup B|\). In other words, there are no wrong predictions (i.e., FP and \(FN = 0\)), and the volumes are perfectly similar. In contrast, JI equating to 0 means that no intersection exists between the ground-truth and prediction, or TP is 0, meaning that the TOI (target of interest) was completely misclassified.
2.2.3 Dice similarity coefficient (DSC)
DSC (or Dice) is the F1 Score counterpart for images, which is a harmonic mean of both precision and recall. In a sense, it measures the similarity between ground-truth set A and generated prediction B. The DSC is defined as
Similar to the JI metric, the two extreme cases are 0 and 1, where the former emphasizes the absence of any similarity and the latter shows the perfect similarity between A and B.
2.2.4 Specificity/true-negative rate (TNR)
As depicted in Eq. (3), specificity investigates the model’s capability in classifying background voxels correctly.
Ranging between 0 and 1, the former denotes a misclassification of all background voxels, and the latter resembles a proper classification of all background voxels.
2.2.5 False-positive rate (FPR)
As shown by Eq. (4), it highlights the amount of error the model is making when classifying background voxels.
Contrary to specificity, a value of 0 is a good indicator of the model’s ability in predicting background voxels. On the other hand, a value of 1 is an extreme scenario where the model wrongly classified all background voxels.
2.2.6 Volumetric overlap error (VOE)
VOE is the complementary metric of JI, which is known as Jaccard distance, knowing that VOE is a special case for volumetric sets. It measures the spatial error represented between the voxels of A and B and is described as
VOE ranges between 0 and 1, where the former means that the voxels of B are perfectly and correctly lying over A’s voxels, and the latter indicates the absence of overlapping voxels between the voxels of A and B.
2.2.7 Average symmetric surface distance (ASSD)
ASSD measures the minimum distance that can be found between a surface voxel in A to another surface voxel in B. Since it is a symmetric metric, the same applies to B with respect to A. Then, the average is taken over all the calculated distances. To define ASSD, we first have to define the minimum distance between an arbitrary voxel v and S(A):
where \(s_A\) is a single surface voxel distance from the surface voxel set S(A).
We can define ASSD as follows:
The value converges to 0 when the highest spatial similarity is achieved. However, the larger the value, the worse the overlap between volumes A and B is noticed, and dissimilarity starts to be observed.
2.2.8 Maximum symmetric surface distance (MSSD)/Hausdorff distance (HD)
MSSD, famously known as HD as well, searches for the maximum distance, defined by Eq. (6), that can be found between volumes A and B.
This metric gives the maximum distance error between A and B, and thus, is extremely sensitive to outliers.
2.3 Datasets
Throughout the last decade and a half, many datasets of different imaging modalities such as CT, contrast-enhanced CT (CE-CT), MRI, and contrast-enhanced MRI (CE-MRI) have been published. Moreover, with the recent development of artificial intelligence (AI) technologies, medical faculty understood how important it is to incorporate such tools to enhance healthcare quality. Thus, datasetsizes have considerably increased. The Segmentation challenge, LIVER Competition 2007 (SLIVER07) happened in a workshop named “3D Segmentation in the Clinic: A Grand Challenge in conjunction with MICCAI 2007. It is considered the first-ever workshop in the liver segmentation field, where it paved the way for the rest of the open-source datasets/challenges, and the results of it are summarized in Heimann et al. (2009). Three years later, 3D Image Reconstruction for Comparison of Algorithm Database (3D-IRCADb) is gathered by the IRCAD institute in France, which includes patients anonymized medical images. In total, the dataset has 22 venous phase CE-CT scans divided into (1) 3D-IRCADb01, which contains 10 males and 10 females with 75% having hepatic tumors; (2) 3D-IRCADb02, which contains 2 CT scans with other abdominal organs segmented. It is worth noting that the majority of literature focuses on the 3D-IRCADb01 group and is normally divided into training and testing records accordingly. In the same year, a very small dataset is also published, called the MIDAS Liver Tumor (MIDAS-LT) Segmentation Dataset. It is a part of a bigger initiative to provide a collection of archived, analyzed, and publicly accessed datasets called MIDAS (Al-Kababji et al. 2023) There have been a few other datasets as well that have been considered by the studies.
3 Segmentation methods
Broadly, there are two types of image segmentation algorithms: (1) discontinuity-based approach—this type of algorithms relies on the abrupt changes (usually at the edge of the objects) in intensity in grey level images. Edge detection is a fundamental tool used in most image processing applications to obtain information from the frames as a precursor step to feature extraction and object segmentation, and (2) similarity-based approach—this type of algorithms group those pixels which are similar in some sense. The task of grouping is performed by the following operations: (a) Thresholding-based operations, (b) knowledge-based operations (including the model-based ones), and (c) region-based operations. All the methods come under these categories; some are either fully automatic or semi-automatic. In this section, we have discussed the automatic and semi-automatic methods to give the reader some sense of methodological notion. The holistic view of the segmentation approaches is provided in Table 4.
3.1 Automatic methods
An automated process should be significantly quicker and require relatively less time to compute, saving both time and money. In the following subsections, we have tried to provide a detailed assessment of each method and the corresponding results.
3.1.1 A model-based validation scheme for organ segmentation in CT scan volumes
Badakhshannoory and Saeedi (2011) present a technique, where pre-computed segmentations of the particular organ is matched with a statistical model. The particular that gives the highest fidelity is considered to be the desired object segmentation. First, a series of segmentations (from under-segmentation to over-segmentation) of a particular data are performed by a general segmentation algorithm. Then determined by principal component analysis (PCA), a statistical model is adapted to produce an organ space. Each candidate’s distance from the organ’s region is measured to determine the candidate producing the best segmentation result. The method was tested on the dataset that contained 30 CT scan volumes (from MICCAI’07). The in-plane resolution for each dataset is \(512 \times 512\) pixels, while the range for inter-slice spacing is from 0.5 to 5.0 mm. On a Computer having an Intel Core 2 Duo (2 GHz) CPU, the segmentation method typically takes around one minute to complete.
3.1.2 Fully automatic segmentations of liver and hepatic tumors from 3-D computed tomography abdominal images: comparative evaluation of two automatic methods
Looking at the error introduced by the operator’s intervention, Casciaro et al. (2011) propose a graph-cut and 3-D initialization method for gradient vector flow (GVF) active contour approach for segmentation. The average intensity of the liver’s statistical model distribution and its standard deviation serve as the foundation for this approach. The original volumetric image is first pre-processed with a mean shift filter to get rid off the noise from homogeneous regions while maintaining distinct and crisp edges. Each slice is partitioned into 64 squares sub-regions; the standard deviation and mean image intensity identify the regions with the most uniform pixel intensity. The liver is symbolized by the median that corresponds to the standard deviation. The dataset consists of 25 anonymised CT individuals, which had voxel sizes ranging from 0.55 to 0.88 \(\hbox {mm}^2\) and a thickness of 2–3 mm. The total time needed to perform the segmentation procedures via graph cut and GVF active contour on a PC with a 3.4 GHz CPU and 1 GB RAM is \(10.9\,\hbox {s} \pm 1.1\) and \(11.5\,\hbox {s} \pm 1.1\), respectively.
3.1.3 Automatic liver segmentation using a statistical shape model with optimal surface detection
Zhang et al. (2010) provide a 3-D generalized Hough transform (GHT) to determine the liver shape model’s approximate position. The model is contorted to modify the liver contour by an ideal graph theory-based surface detection after statistical shape model (SSM) adaptation. Some preprocessing operations such as edge detection, down-sampling, and smoothing of the input image are performed before the actual procedure is adapted. It is a 4-step process, viz. (1) Shape model construction, (2) 3-D GHT liver localization, (3) SSM subspace initialization, and (4) Graph theory based optimal surface detection approach. The datasets used in this study were from MICCAI 2007 Grand Challenge. For the building of shape-models, 40 additional CT volumes with normal liver architecture were employed. Each model has 5120 triangles and 2562 evenly distributed vertices. The segmentation procedure is finished in \(4.47\,\hbox {s}\) on a 32-bit computer (2.33 GHz Core 2 and 2 GB RAM).
3.1.4 Automatic segmentation of the liver from multi- and single-phase
This article by Rusko et al. (2009) proposes a region growing technique that is independent of the acquisition process. The algorithm is aimed to segment single and multi-phase CT images. An initial segmentation is constructed in the first stage employing all phases. Each phase is subjected to the three processes that follow independently. The first segmentation results and the original input images serve as the input for these phases. The steps are: (a) selection of seed region in the liver, (b) method of region growing for segmentation of liver, and (c) post processing. The seeds region is selected assuming an empirical value for liver region intensity. The results are registered once the segmentation is available for each step, allowing the computation of the ultimate result as a sum of all stages. An automated contrast-enhanced CT scan takes \(25.6\,\hbox {s} \pm 7.2\) to process on average utilizing an Intel Core2 Duo processor at 2.2 GHz CPU and 2 GB of Memory. The segmentation for single phase takes \(40.7\,\hbox {s} \pm 9.4\) to complete the process; MICCAI 2007 training dataset is used in this study.
3.1.5 A new fully automatic and robust algorithm for fast segmentation of liver tissue and tumors from CT scans
Massoptier and Casciaro (2008) present a 3-D fully automated model-based method that relies on statistical information of images. It is a 3-step procedure, viz. (1) pre-processing—the noise from homogeneous regions is eliminated from the original volume image using a 3-D mean shift filter, (2) liver-specific statistical model discrimination—the aim is to identify the most liver representative area in the volume dataset. The volume that is pre-processed is split into 64 squares portions, and for each slice, the standard deviation and mean image intensity are calculated. Then, for all volume slices, the internal regions that have the least standard deviations are separated out and arranged in descending order of mean values. Then, the liver is linked to the vast majority of those organs, and (3) liver surface segmentation refinement—finally, GVF active contour method is applied to obtain the liver surface. Twenty one distinct patient CT datasets are employed in this experiment. The slice thickness, pixel size range, and imaging matrix are from 0.55 to 0.88 \(\hbox {mm}^2\), 2–3 mm and \(512 \times 512\), respectively. On a personal notebook with 3.4 GHz and 1 GB memory, on average the processing time for a single slice is \(11.4\,\hbox {s} \pm 1.2\).
3.1.6 Construction of a probabilistic atlas for automated liver segmentation in non-contrast torso CT images
Zhou et al. (2005) suggest a technique based on diaphragm warping to normalize the liver’s proper anatomical position. Subsequently, a probabilistic atlas is constructed for liver segmentation from CT images of non-contrast torso. It is a 3-step algorithm, viz. (1) likelihood of liver region is calculated, (2) the liver is normalized using warping of diaphragm and thin plate spline method (Bookstein 1989) and after that, a liver image is created by a great deal of pre-segmented liver areas projected into three dimensions. The liver’s density distribution is then approximated using a Gaussian model. Gaussian parameters are determined by measuring the region’s density histogram so that each voxel satisfies a probability criterion, and (3) liver segmentation performed using the atlas. This study uses a total of 80 CT scans having non-contrast torso patient cases. Each image has 12 bits of resolution and 0.6 mm of spatial resolution.
3.1.7 Automatic liver segmentation for volume measurement in CT images
Lim and Ho (2005) suggest a method using the prior knowledge to find the consistent areas that belong to liver. This is a 3-step process, viz. (1) image simplification—the region of interest, ROI, is decided by dividing the abdomen CT image into \(64 \times 64\) pixel blocks and then discarding the unnecessary blocks. To make the liver appear significant, a multilevel thresholding is used, (2) search range detection—the low order multiscale morphological operations are performed on the thresholded image to locate the first and the second search areas. Many scales of morphological filtering are recursively performed to get the primary and the subsidiary search areas. The terminal search area is obtained by eliminating the subsidiary area from the primary search area. A modified K-means algorithm succeeded by a morphological analysis operation are implemented to detect the fine liver areas. The dataset consists of 10 patients, and the samples are contrast-enhanced venous phase CT scans with a 5 mm spacing and \(512 \times 512\). On a Pentium 4 3.0 GHz processor, total processing time typically ranges between 1 and 3 min per slice.
3.1.8 Construction of an abdominal probabilistic atlas and its application in segmentation
Park et al. (2003) propose an unsupervised segmentation with maximization a posteriori probability (MAP) and probabilistic atlas. It is 2-step method, viz. (1) atlas construction—the individual dataset registration onto target reference is implemented with a similarity measure, mutual information (MI). The unnecessary compressible organs require warping transform like thin plate spline (TPS) for the same. A decent registration accuracy is attained by registering each organ separately, and (2) liver segmentation—if the observed data and probabilistic atlas are indicated by \({{\textbf {Y}}}\) and \({{\textbf {A}}}\), respectively, the problem lies to find the true label field \({{\textbf {X}}}\). For this, a cost function MAP is defined and the probabilities of \({{\textbf {Y}}}\) are Gaussian modeled. The standard consequence for nearby dissimilar objects is included as Markov random field (MRF) priors. Iterated conditional mode (ICM) (Besag 1986) is used to optimize the posterior probability in the MRF set-up. The segmentation technique is tested on 20 abdominal CT data with slice thicknesses ranging from 7 to 10 mm.
3.1.9 Liver segmentation from computed tomography scans: a new algorithm
Campadelli and Esposito (2009) present a gray level methodology to automatically extract the liver samples and segment using \(\alpha\)-expansion algorithm (Boykov and Kolmogorov 2004). It is a 3-step method, viz. (1) heart-liver separation—the largest linked area that connects the bounding box in the image is thresholded, (2) gray levels estimation of the liver—on a liver sample set, a 3-D box below the heart typically defines the liver tissue. Again, the 3-D body box (patient’s body) is divided by the alpha-expansion algorithm with the aid of the graph-cut approach into five groups (liver, spleen, bones and kidneys, stomach, and organs with comparable gray levels background) (Kolmogorov and Zabih 2004). This method cycles over the five labels in a random order to determine a binary evaluation for each label. The liver is the organ with the largest volume and lowest label, and (3) liver volume refinement—this action is necessary to eliminate the unwanted parts in the liver. Around 40 abdominal contrast-enhanced CT data from the third phase are used to assess this strategy. Each slice has a resolution of \(0.625\,\hbox {mm} \times 0.625\,\hbox {mm}\) and a pixel size of \(256 \times 256\) for a total of around 80 axial slices with a 3 mm spacing for each patient. When using a Pentium IV processor operating at 3.2 GHz, the method completes the task in less than 50 s.
3.1.10 Patient-oriented and robust automatic liver segmentation for pre-evaluation of liver transplantation
Selver et al. (2008) propose a patient oriented 3-step segmentation method, viz. (1) pre-processing to remove the irrelevant tissues and find ROI (liver) from the primary image. The volumetric histogram of the input image is subjected to an adaptive thresholding to find and delete the lobes corresponding to the irrelevant tissues, (2) classification of liver—a modular classifier consisting of K-means and multi-layer perceptron (MLP) network is used to segment liver starting from the first through end slice in an iterative manner. The initial image is chosen around one third of the series and prepared using Ostu’s method (Otsu 1979) to separate the unwanted muscle tissues (dark organs) keeping the liver, spleen and heart (brighter organs), and (3) post processing—this action is necessary to eliminate small pseudo segmented objects. A total of 20 data sets of 12-bit DICOM images with a slice thickness of 3–3.2 mm and a resolution of \(512 \times 512\) are utilized in this investigation. The K-means algorithm application’s Java variant runs approximately 12–17 min on a typical Computer with 2 GB of RAM and a 3 GHz CPU. K-means classifier is used in the Matlab version, which takes around 30 min. Both in Matlab and Java, the MLP classification method is completed within 45 min.
3.1.11 Fully automatic anatomical, pathological, and functional segmentation from CT scans for hepatic surgery
Soler et al. (2001) propose an anatomical segmentation technique built on the conversion of topological, geometrical, and morphological constraints from anatomical knowledge. Just before characterizing the bones, the image tangential tissues are enhanced with proper thresholding, followed by morphological operation. The range of intensity of the kidneys, spleen, and liver parenchyma, which are equally located on both histograms, may be discovered by comparing the gray-level histograms. By executing a thresholding followed by morphological operators, the kidneys and spleen are distinguished. The liver is then extracted using the Montagnat and Delingette approach (Montagnat and Delingette 1996), which treats the global transformations calculated in the registration framework (Brown 1994) as a deformation field. The locality parameter and combined force of this approach are applied to each model vertex. As a way to define the liver, the framework adds a global restriction to the deformation process. A total of 33 intravenous injection data and two portodata build the database for this study. A collection of 35 CT data with thickness ranging from 2 to 3 mm are used for the experiment.
3.1.12 Automated segmentation of the liver from 3D CT images using probabilistic atlas and multilevel statistical shape model
Okada Yokota et al. (2008) discuss a method that uses two groups of atlases, i.e. the probabilistic atlas (PA) and the statistical shape model (SSM). Spatial standardization of input data (by radiologist) gives average liver shape in the set. Each patient dataset is converted into the systemized patient space through nonrigid registration. PA is constructed by the average of binary images specifying 1 for liver and 0 for the rest in all patient datasets. To build multi level SSM (MLSSM), a multiple-level surface model is first constructed by dividing a liver shape into patches. The patches are then recursively divided to form MLSSM. The liver segmentation is performed in 3 steps: (1) the spatially standardized CT data is smoothed by anisotropic diffusion filtering. The volume of interest corresponds to the area, where PA surpasses a certain amount of threshold, (2) the initial shape parameter is determined from surface model produced from the initial area by the reduction of a cost function to accommodate the Euclidean distance between a point and a surface, and (3) segmentation—analysis of the CT volumes along the MLSSM surface normal yields the edge points of the liver borders. In this experiment, 28 abdominal CT datasets (159 slices, pitch: 1.25 mm, slice thickness: 2.5 mm, \(512 \times 512\) matrix) are used.
3.1.13 Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation
Linguraru et al. (2010) [extension of Linguraru et al. (2009)] suggest a method based on PA to segment liver. The algorithm works in 2 stages as (1) atlas construction—the reference image (R) is chosen at random from the input database, while the other images are designated as I. The images I are re-scaled and registered to R organ-wise based on normalized MI (Studholme and Hawkes 1999). The registered livers are translated in the atlas based on the average normalized centroid, the probabilistic organ atlases are then computed. From this step, those models are extracted that are conservative (A), and (2) liver segmentation— the spatial normalization is then applied to both A and barA after performing a global affine registration between R (from the atlas creation) and I. A more flexible alignment is needed to offset the remaining deformation \({\bar{A}}_r\) with the use of B-splines (Rueckert 1999). The registration provides a preliminary estimate of the target organ. A geodesic active contour (GAC) (Caselles and Sapiro 1997) is implemented to accommodate for potentially missing liver sections. Ten abdomen non-contrast CT data without anomalies were utilized to create the probabilistic atlas. With inter-slice spacing of 1 mm, the image resolution ranges from 0.54 to 0.77 mm. For segmentation of livers, 257 abdominal CT scans are used with image resolution and inter-slice distance as from 0.62 to 0.93 mm and from 1 to 5 mm, respectively.
3.1.14 A deformable model for automatic CT liver extraction
Gao and Kak (2005) present a Spedge-and-Medge-based algorithm for liver delineation. This is a 2-step method, viz. (1) coarse segmentation—a \(5 \times 5\) median filter on the original image reduces the impulse noise present in it. The Canny edge operator produces the primitive object regions and the edge image is subjected to a modified split-merge method (Spedge-and-Medge) separating out the coherent areas. The liver areas are calculated using geometric and non-geometric properties. After then, the areas are combined to form a single and sizable region, (2) refinement of boundary—the rough border acquired in step 1 is smoothed using a modified active contour model. The method builds chords to each of the successive boundary points under a certain threshold criterion, starting at a boundary point. Finally, the energy minimization of the contour at the boundary consisting of the control points is performed to get the desired contour. In this experiment, 15 patient data were used with 5 mm collimation and \(512 \times 512\) image resolution.
3.1.15 Cognition network technology for a fully automated 3D segmentation of liver tumors
The effective context-based methodology is proposed by Schmidt et al. (2007), where the liver is split mechanically based on its anatomical location. The following two components are the key in the algorithm. The heuristic threshold values for the intensity are used to partition the liver in the 3D data set. Depending on their intensity and volume, the image objects are further improved to approximate additional unneeded bodily organs. These body components serve as the foundation for the calculation of a new layer of 3D edge data, which ultimately serves as a guide for additionally perfecting the body parts. The liver, which lies below the right lung and is bounded by the skeleton and gall bladder, is presented as the image object with the greatest volume. Ten datasets were included in this study. Using a machine with a two core CPU (2.4 GHz, 3.5 GB RAM), the procedure takes from 3 min (for a data set with 145 slices) to 10 min (for a data set with 304 slices) to finish the operation.
3.1.16 Fully automatic segmentation of liver and hepatic tumors from 3-D computed tomography abdominal images: comparative evaluation of two automatic methods
Casciaro et al. (2012) develop a method combining graph cut and active contour algorithms with gradient flow. The method is tested on 52 patient data and the segmentation accuracy is evaluated using False-Positive Rate (FPR), False-Negative Rate (FNR), Dice Similarity Coefficient (DSC); they are found to be 2.39%, 5.10%, and 95.49%, respectively.
3.1.17 Modeling n-furcated liver vessels from a 3-D segmented volume using hole-making and subdivision methods
Yuan et al. (2011) propose a method for modelling n-fork subtrees, where cross-sectional contours and vessel are extracted from centre-line. A polygonal mesh with cross-sectional contours is then constructed for each branch in descending order. The experimental results show that smooth mesh models could be generated automatically for n-branch vasculature with an absolute error of 0.92 (average) voxels and an average relative error of 0.17.
3.1.18 Medical image segmentation by combining graph cuts and oriented active appearance models
Chen et al. (2012) combine Graph Sections (GC) and Live Wire (LW) with the Active Appearance Model (AAM). Using the GC parameters and LW cost function, AAM is generated and trained during the model building stage. It incorporates AAM and LW to produce an oriented AAM (OAAM). The multi-object OAAM mechanism is used to slice the organs using an adapted pseudo-3D strategy. The iterative GC-OAAM is used to mark the objects. The method is tested on MICCAI 2007 liver data; the pseudo-3D OAAM method performs similar to the conventional 3-D AAM method while running 12 times faster.
3.1.19 ACM-based automatic liver segmentation from 3-D CT images by combining multiple atlases and improved mean-shift techniques
Ji et al. (2013) present a liver segmentation algorithm based on automatic context model (ACM), which combines ACM, multi-atlas, and mean transfer techniques. It is a two-step learning-based method; in the initial training step, ACM-based classifiers with multiple ranks are utilized, the test image is then segmented in each space of the atlas using each sequence of ACM-based classifiers. Using a multi-class fusion technique, the results of all atlas space segmentation are combined to produce the final result. The data from the MICCAI 2007 are used to evaluate the proposed method. The method has claimed to have significantly reduced the segmentation time from approximately 400 to 35 min by introducing region-based labelling and employing an improved mean-shift algorithm. The entire segmentation process is less than one hour.
3.1.20 Automated abdominal multi-organ segmentation with subject-specific atlas generation
Wolz et al. (2013) suggest a multi-organ abdominal segmentation using atlas-based method. This is claimed to have applied to multiple organs without changing specialization and individual parameters. The atlas registration and a weighting system are used to subject-based priorities from an atlas database by combining a patch-based segmentation and multi-atlas registration. The final segmentation is then generated using the automatically learned intensity model in the graph cut optimization phase, which contains high-level spatial information. The segmentation method is evaluated on 150 CT data. The values of overlap of DSC for liver, kidney, pancreas, and spleen are found to be 94%, 93%, 70%, and 92%, respectively.
3.1.21 Joint probabilistic model of shape and intensity for multiple abdominal organ segmentation from volumetric CT images
Li et al. (2012) develop a joint probabilistic model determining a probability map, when a voxel belongs to specified object with an estimated shape. Probabilistic principal component analysis is used to explain the shape variation and reduce computational complexity using expectation maximization. 72 CT training datasets are used to create shape models of the liver, spleen, and kidney. To highlight 3D visualization colour coding is used. The algorithm was evaluated on 40 test datasets that were divided into normal (34 normal cases) and pathologic (six datasets) classes.
3.1.22 Automatic liver segmentation based on shape constraints and deformable graph cut in CT images
Li et al. (2015) suggest a framework comprising three steps: (1) data processing; (2) initialization parameters; and (3) data segmentation. Initial principal component analysis-based statistical shape models are created, and a filter is used to smooth the input image. Then, the mesh is locally and iteratively transformed into a boundary-constrained mesh to remain close to the shape subspace, and the average shape model is altered through thresholding and Euclidean distance to obtain an approximate location on the image. Finally, graph cut integrates the features effectively and relationships of the input images and the initialized surface for accurate liver surface detection. In this method, 50 CT data are used from two databases, 3Dircadb and liver07. The proposed method took approximately 5 min and 3 min to compute the segmentation on the Sliver07 database and 3Dircadb database, respectively.
3.1.23 An improved confidence connected liver segmentation method based on three views of CT images
Song et al. (2019) propose a fusion of segmentation of liver that combines liver segmentation results from three perspectives. An advanced curved anisotropic diffusion filter is first used to reduce noise, which records edge information simultaneously. Second, liver intensity statistics and analysis are automatically used to select liver seed points. With the method based on reliable association, the contours of the liver are extracted from three views of the CT image, and the cavity filling method is used to improve the contours. Finally, they combine coronal, sagittal, and cross-sectional views of the liver. Ten abdominal CT data are used for clinical validation. The method achieves a Dice score of 97.
3.1.24 Towards liver segmentation in the wild via contrastive distillation
Fogarollo et al. (2023) propose a contrastive distillation scheme using a pre-trained large neural network to train their model that is reported to be small. They first extract the features by a self-supervised Vision Transformer (ViT) and then carry out contrastive distillation on the obtained features. They map the neighboring slices close together in the latent representation, while mapping distant slices far away. They use ground-truth labels to learn a U-Net style upsampling path and recover the segmentation map. The method is evaluated and compared on different medical datasets such as well-known BTCV, CHAOS, IRCADb, LiTS, ACT-1K, and AMOS221. They obtain an average Dice score, ASSD, and MSSD as \(0.918 \pm 0.066\), 1.3 mm, and 5 cm, respectively.
3.1.25 Automatic 3D CT liver segmentation based on fast global minimization of probabilistic active contour
Jin et al. (2023) propose a liver segmentation method based on a probabilistic active contour (PAC) model and its fast global minimization scheme (3D-FGMPAC), which is reported as explainable as compared with deep learning methods. A slice-indexed-histogram is initially constructed to localize the volume of interest estimating the probability that a voxel belongs to the liver according its intensity. The 3D PAC model is initialized using the probabilistic image. The combination of gradient-based edge detection and Hessian-matrix-based surface detection is used to produce a contour indicator function. The initial probabilistic image contour is then evolved into the global minimizer of the model by a fast numerical scheme showing a smoothed and highlighted probabilistic liver image. Finally, a region growing method is applied to extract the liver mask. After testing the method on two public datasets, the average Dice score, volume overlap error, volume difference, symmetric surface distance and volume processing time are found to be 0.96, 7.35%, 0.02%, 1.17 mm and 19.8 s for the Sliver07 dataset, and 0.95, 8.89%, \(-\)0.02%, 1.45 mm and 23.08 s for the 3Dircadb dataset, respectively.
3.2 Semi-automatic methods
Semi-automatic methods are usually considered less accurate in comparison to the automatic ones, because the operator intervenes in the due course of getting the segmentation output and they are prone to error. However, these methods have also merits as discussed below.
3.2.1 Advanced fuzzy cellular neural network: application to CT liver images
A novel fuzzy cellular neural network (AFCNN) is proposed by Wang et al. (2007) that primarily addressees the problem, when the liver’s CT imaging borders overlap with those of other organs. AFCNN retains the feed-forward and feedback stimuli, but the cell status with regards to its neighbor cells is used in place of the fuzzy feed-forward and feedback stimuli. With the help of this technique, AFCNN uses both liver and the non-liver region, improving the segmentation accuracy. The boundary line in the segmented liver area tends to be smoother as the number of FCNN iterations increases, making it less probable to reproduce the original liver boundary. In this experiment, five CT datasets are used on a computer with 128 MB of memory and Matlab 6.5.
3.2.2 A knowledge-based technique for liver segmentation in CT data
Foruzana et al. (2009b) present a slice-based liver segmentation method that typically generates the liver mask by connecting the bones. It begins from the initial slice and runs past all slices sequentially. The first slice is chosen to match to the large cross section area. By thresholding in the range (400, 1700) HU, the bones are located. The bone mask in the current slice is estimated using the connection of the ribs in the previous slice. The liver areas are split into a number of smaller regions by utilizing split thresholding. The boundary of liver is performed by dividing the whole range into two overlapping ranges and a decision is made with the help of morphological criteria, whether each range is part of the liver or not. There are 50 subjects and each data set contains 157–279 images with 12 bit DICOM images. Using a Windows Computer with a P4 (3 GHz) and 2 GB of memory, the technique needs roughly 8 min to segment a dataset with 160 slices.
3.2.3 Liver segmentation for CT images using GVF snake
Liu et al. develop a method for removing concavities from aberrant livers, particularly those with lesions at the liver edges (Liu et al. 2005). It is a three-step process, beginning with the estimation of a primary edge map. Two thresholds are set, one on either side of the peak of the liver in the histogram of the median filtered image as a guide. (2) GVF field and starting contours—the computation of GVF field is conducted on the edge map. The liver is identified as the greatest volume between these two thresholds. The potential initial and empty region contour (of the GVF field) are both taken into account when determining the initial liver contour. Next, the initial contour is refined by the GVF snake to determine the liver boundary. 20 contrast-enhanced volumetric liver data with many big dispersed lesions are used in this study. Also, 551 two-dimensional liver images from 20 patients are taken into account, each showing colorectal metastases that have spread throughout the livers. The dimensions of all slices are \(512 \times 512\) pixels, in-plane pixel size range is \(0.56 \times 0.56\,\hbox {mm}^2\)–\(0.87 \times 0.87\,\hbox {mm}^2\), and the slice thickness is 3.75, 5.0, 7.0, and 7.5 mm, respectively.
3.2.4 Computer-aided measurement of liver volumes in CT by means of geodesic active contour segmentation coupled with level-set algorithms
The method given by Suzuki et al. (2010) presents an approach based on GAC coupled with a level-set algorithm to segment liver in hepatic CT. (1) pre-processing—anisotropic diffusion algorithm (Perona and Malik 1990) is used in the first part of this two-step process to minimize noise, maintain structures, and enhance anatomical structures in the input portal-venous-phase CT images. (2) Liver extraction: the fast marching (FM) level-set algorithm (Sethian 1996) is utilized to calculate an irregular liver contour. The FM level-set algorithm describes the advancement of a closed contour (or curve) as a function of time and speed in the normal direction at a specific place on the contour. The seeding points within the hepatic region needed by the FM algorithm are provided by the radiologist. As a result, the anatomical boundaries of the liver can be expanded using the FM algorithm. To get a close approximation of the liver border, the GAC refines the initial contour provided by the FM method. The database comprises 15 patients with reconstruction intervals of 2.5 mm or 3 mm and collimation of 3 mm or 4 mm. Each of the reconstructed slices of CT features a \(512 \times 512\) matrix with pixels ranging from 0.5 to 0.8 mm in size. The approach takes 2–5 min to perform on a Computer (Intel, Xenon, 2.7 GHz).
3.2.5 Liver segmentation by intensity analysis and anatomical information in multi-slice CT images
Foruzan et al. (2009a) present a 5-step method, viz. (1) manual liver segmentation—the largest middle slice of the liver is first found out manually and this slice is the starting point in the segmentation process, (2) estimation of liver intensity range—to estimate the statistical properties of the liver, a Gaussian mixture model with two components is used, since the histogram of a segmented liver in a single slice is composed of two Gaussian distributions, (3) ROI for liver is determined by segmenting out the ribs, (4) heart is separated from liver by simple thresholding, and 5) thresholding a slice—the liver’s histogram divides the entire intensity range into two categories: lower range and higher range. Next, local analysis chooses several threshold values for each location to categorize liver and non-liver tissues. A data with dimensions of \(512 \times 512 \times\) and 150 slices is segmented in its whole on a PC (P4 CPU, 2 GB) within 6 min. This study has used the MICCAI 2007 Grand Challenge data for the experiment.
3.2.6 An entropy-based multi-thresholding method for semi-automatic segmentation of liver tumors
The watershed technique (Choudhary et al. 2008) is used to extract the liver contours, and a minimal cross-entropy multi-thresholding approach is used to segment the tumors. The stages are (1) Simple thresholding and shape-preserving Cubic-Hermite interpolation are used to segment the ribs on all the slices. Moreover, these curves serve as restrictions for the segmentation of the liver. (2) Diaphragm segmentation: to limit the area to the liver alone, a diaphragm location method is used (Beichel et al. 2002). The program calculates the gradient magnitude of the selected slice, analyzes the data to highlight borders, and then chooses the slice, where the liver is prominent. The watershed transform (Vincent and Soille 1991) is then applied if the user-defined point is located inside the liver. The class that contains that point is the liver, and (4) segmentation of tumors—slice after slice, a minimal cross-entropy multi-thresholding method (Li and Lee 1993) is used to segment the tumors. 10 CT datasets of cancer patients were used.
3.2.7 A 3-D liver segmentation method with parallel computing for selective internal radiation therapy
Goryawala et al. (2011) describe a method for 3-D liver segmentation that consists of a modified k-means segmentation method and a local contour algorithm. In this method, five distinct locations are identified in the CT scan frames during the segmentation process. This paper provides advantages of developing parallel computing-aware algorithms in medical imaging prior to investing in a very large-scale distributed system. The algorithm is independent of dataset characteristics such as liver structure, size, location, and distribution of intensity. The results from single workstation show a 78% reduction in calculation time from 4.5 h to near about 1 h. The accuracy after calculating the volumes of the liver and tumor area is reported to have an average error of less than 2%. Experiments with up to 2 slices are used to evaluate the effect of parallelism.
3.2.8 A likelihood and local constraint level-set model for liver tumor segmentation from CT volumes
Li et al. (2013) describe a level-set based model that combines the edge energy and probability. With the density distribution of multimodal in the background, which may contain multiple regions, probabilistic energy minimization estimates the distribution of density of the damaged part. The edge detector keeps the ramp associated with the boundaries in the edge energy composition for weak boundaries. The Chan-Vese and geodesic plane series models, in addition to clinician-manual segmentation, are compared to this approach. The suggested model outperforms the geodesic plane model in liver tumor segmentation, where the Chan-Vese model was reported to be unsuccessful. The liver RVD (relative volume difference), JDE (Jaccard distance error), ASD (average surface distance), SD (surface distance—RMS type), and the SD (surface distance-maximum) are \(8.1 \pm 2.1\) percent, \(14.4 \pm 5.3\) percent, \(2.4 \pm 0.8\) mm, \(2.9 \pm 0.7\) mm, and \(7.2 \pm 3.1\) mm, respectively.
3.2.9 An efficient and clinical-oriented 3D liver segmentation method
With this technique, the segment of liver is automatically divided by the portal vein branches (Zhang et al. 2017). The regulation of the branches of the portal vein is based on artificial segmentation taking into account the distribution of the vessels. 20 CT image datasets from the MICCAI 2007 liver segmentation challenge and 60 datasets from Zhu Jiang Hospital of Southern Medical University were used in this experiment. The mean VOE, RVD, and RMSD are 7.76%, 3.44%, and 2.81%, respectively.
3.2.10 Liver segmentation on CT and MRI using Laplacian mesh optimization
Using MRI and CT scan images, Gabriel Chartrand et al. (2016) develop a semi-automated liver segmentation method. A crude 3D model of the liver is first created using some user-generated contours to broadly outline the shape of the liver. A Laplacian network optimization method is then used to autonomously modify the model until the patient’s liver is well defined. A correction tool is introduced that allows the user to modify the segmentation. The method is tested on SLIVER07 dataset. The mean volume overlap error is 5.1% with a mean segmentation time of 6 min.
3.2.11 Semi-automatic liver segmentation based on probabilistic models and anatomical constraints
Le et al. (2021) present a graph-cut based method for multivariable normal distribution of liver tissues. An internal patch is used to construct a subject-specific probability prototype using a user-specified seed point. Then, an iterative assignment of pixel labels is used to gradually update the spatio-contextual data-based probabilistic map of the tissues. The graph-cut model is then optimized in order to extract the 3-D liver. On the SLIVER07 dataset, the system was tested. In all, 25 asymptomatic and 2 symptomatic cases were examined. Because of trivial assertions about anatomical and geometrical structures, the entire procedure only lasted 1.3 min to segment a complete 3D liver; furthermore, the values of VOE, RVD, ASD (or ASSD), RMSD, and MSD (or MSSD) were \(8.0\pm 1.1\), \(0.3\pm 2.7\), \(1.3\pm 0.4\), \(2.5\pm 1.0\), and \(24.9\pm 10.0\), respectively.
3.2.12 Towards liver segmentation in the wild via contrastive distillation
Fogarollo et al. (2023) propose a contrastive distillation scheme using a pre-trained large neural network to train their model that is reported to be small. They first extract the features by a self-supervised Vision Transformer (ViT) and then carry out contrastive distillation on the obtained features. They map the neighboring slices close together in the latent representation, while mapping distant slices far away. They use ground-truth labels to learn a U-Net style upsampling path and recover the segmentation map. The method is evaluated and compared on different medical datasets such as well-known BTCV, CHAOS, IRCADb, LiTS, ACT-1K, and AMOS221. They obtain an average DICE score, ASSD, and MSSD as \(0.918 \pm 0.066\), 1.3 mm, and 5 cm, respectively.
3.2.13 Towards liver segmentation in the wild via contrastive distillation
Jin et al. (2023) propose a liver segmentation method based on a probabilistic active contour (PAC) model and its fast global minimization scheme (3D-FGMPAC), which is reported as explainable as compared with deep learning methods. A slice-indexed-histogram is initially constructed to localize the volume of interest estimating the probability that a voxel belongs to the liver according its intensity. The 3D PAC model is initialized using the probabilistic image. The combination of gradient-based edge detection and Hessian-matrix-based surface detection is used to produce a contour indicator function. The initial probabilistic image contour is then evolved into the global minimizer of the model by a fast numerical scheme showing a smoothed and highlighted probabilistic liver image. Finally, a region growing method is applied to extract the liver mask. After testing the method on two public datasets, the average Dice score, volume overlap error, volume difference, symmetric surface distance and volume processing time are found to be 0.96, 7.35%, 0.02%, 1.17 mm and 19.8 s for the Sliver07 dataset, and 0.95, 8.89%, \(-\)0.02%, 1.45 mm and 23.08 s for the 3Dircadb dataset, respectively.
4 Summary of results
The methods are summarized in Tables 2, 3, and 4. The papers that have provided the complete quantitative results are included in these tables; the name of the contributors, type of method, data used, its advantages, execution time, and the performance quality are included. The situation for which a method is suitable clinically has been highlighted in Table 5. It may also be noted that we find difficulty as some papers provide insufficient information in their evaluation process.
5 Discussion
We discuss the potential drawbacks of the methods, both technically and clinically.
5.1 Technical analysis of each method
Model-based methods (Badakhshannoory and Saeedi 2011; Massoptier and Casciaro 2008; Soler et al. 2001) seem accurate, but the construction of a robust model seems to be a tough task with respect to the liver shape variation. Moreover, sufficient training datasets are needed to build the model. The technique reported in Soler et al. (2001) needs larger medical validation to evaluate the algorithm’s durability. Although the algorithm (Massoptier and Casciaro 2008) is reported to have required less time as compared to the other model-based methods, the amount of learning data required seems to be still high. Campadelli and Esposito (2009) suggest a technique based on gray levels that combines with \(\alpha\)-expansion algorithm and graph-cut method for liver segmentation. Unfortunately, a major drawback associated with \(\alpha\)-expansion is its linear time complexity. Moreover, it is well known that graph cut is sensitive to noisy data, therefore, its robustness is a genuine concern. Furthermore, it relies on the primary image for correct seed placement. Thus, if the user does not choose an appropriate initial slice, one may not get an accurate final output. Zhang et al. (2010) propose a statistical shape-based contour extraction method, however, the manual demarcation of the training set is time consuming and poses a practical restriction in the model’s construction. Again, GHT has some limitations, viz. (1) to detect the bin amid the high background noise (which is inherent in medical images), high number of votes must have to fall in the bin, (2) if the quantity of parameters (m) is larger, less number of votes would fall in a bin. So the bins corresponding to the real figure are sometimes not appeared, and the complexity increases with number of parameters.
Rusko et al. (2009) present a region growing-based method, which is quick and easy. However, the approach has certain limitations, especially, when there is either high amount of noise or the segmentation region’s intensity is not consistent. further, it lacks in fixing the problem of spatial correlation to get a coarse contour. Zhou et al. (2005), Okada Yokota et al. (2008) and Linguraru et al. (2010) propose methods based on probabilistic atlas, but they are computationally expensive and the subsequent problems due to this are well known (Zhai et al. 2019). Additionally, the exact alignment of the atlases with the target image is crucial for the algorithms’ accuracy. The pairwise registration, which is frequently utilized, may provide erroneous alignment, particularly, across images with significant changes. Simultaneously, it is difficult to describe a proper joint probability function. It is reported by Whiteley et al. (1998) that the distribution of local standard deviation values in the input image determines the distribution after local mean removal. A concern in almost every method (Okada Yokota et al. 2008) is its validation; most of the methods are tested on a smaller dataset. Since TPS generates quantitative analyses of spatial organizational shapes with varying sizes in terms of features and localizations, Park et al. (2003) present a method utilizing TPS to suppress unwanted tissues. However, TPS includes curvature discontinuity at the identical experimental surface data points, which further complicates in estimating the change.
Foruzan et al. (2009a) suggest a method using expectation maximization (EM), but the initial parameters of EM should be properly chosen since it influences the performance of the algorithm. EM is conceptually simple, easy to implement, and at each iteration, marginal log-likelihood is improved. However, after a few steps, the rate of convergence becomes excruciatingly slow as one approaches a local optima. The proposed GVF snake-based method by Liu et al. (2005) is efficient in solving initialization issues and a lackluster convergence to border concavities present in liver images. However, it is little suitable for boundaries in noisy images, where the shape of edges are zig-zag in nature. Linguraru et al. (2009) suggest an autonomous, atlas-based technique, however, care must be taken while designing the atlas. The problem appears, when an atlas designed for one image having no tumor is used for another image having multiple tumors. Moreover, the utility of atlas-based segmentation has been restricted in the existence of significant space-consuming lesions, where it tends to deform and shift liver structure during the registration. This type of CT images resemble to Budd–Chiari Syndrome (Arora et al. 1991). Schmidt et al. (2008) suggest a cognitive network language-based technique, driven by knowledge, has significant flaws. For instance, the region growing algorithm fails to expand into smaller liver lobes, and the big tumors that largely cover the liver.
The discontinuous input–output mapping in the FCNN (Wang et al. 2007) is a limitation in the learning phase. The method by Selver et al. (2008) contains a switching mechanism that depends on the contrast level of the tissues. In this method, the gradient descent may reach a local minima instead of the global minima since the objective function of the network in an MLP-based mechanism and it is not necessarily convex. The conventional snake (Gao and Kak 2005) is quite popular due to its simplicity; however, the contours usually become trapped onto false image features, and in terms of extracting non-convex features. It is well known that method proposed by Canny is a multi-step edge detection procedure, which detects the edges suppressing the noise. Therefore, noise removal in the preprocessing step is not necessary, because excessive smoothing (without edge preserving characteristics) of the original image also smooths the edges. Level set (Suzuki et al. 2010) has been used extensively, because of the method’s ability to extract curved objects with complicated topology, resistance to noisy, and clear numerical framework of multidimensional implementation. However, the calculation time needs to be significantly lowered for the approach to be viable in clinical application. Additionally, care must be taken while empirically assigning values to the parameters and deciding a suitable speed function. Knowledge-based image segmentation methods (Foruzana et al. 2009b; Lim and Ho 2005) maximize the posterior probability across the space domain and divide the image domain compromising between data attraction and shape fit with the prior model. Knowledge of the location, shape, and gray level of the liver is important and crucial to predict these information a priori (Lim and Ho 2005). It is usually difficult to always probe the shape of the region properly due to filter’s bunt shape. The approaches might underperform on large, diverse and complicated datasets. Choudhary et al. (2008) provide an entropy-based multi-thresholding approach that employs watershed segmentation and cross-entropy multi-thresholding to extract the organ boundaries. While the segmentation is accurate in higher for smaller and uniform tumors, it does not provide a good accuracy for larger ones, mostly because of the increased non-uniformity in their intensities.
5.2 Outlook
In Sects. 3, 4, and 5, we discussed all the popular automated and semi-automatic methods along with their advantages and disadvantages. Each technique accomplishes a defined goal. Automatic methods are generally preferred, however, suitable training dataset (with respect to size and quality) is required to develop them. Sometimes due to larger datasets, some automatic methods need more time than desired. The datasets used in all these methods for training and testing are very limited in number, therefore, there has been no “one for all” method. There is another problem: although the clinicians prefer to use automatic methods, sometimes they prefer to correct the output based on their requirements. In this scenario, the automatic methods pose challenges, because they usually do not allow. On the other hand, the semi-automatic methods are simple and relatively easy to implement. However, these approaches are usually operator dependent, thus, has greater chance to introduce larger divergence in the algorithm performance. The semi-automatic methods are probably more suitable for clinical practices than the automatic ones, if they can cover the issues owing to the large levels of inter-personal and intra-personal variability. If all the methods are examined sequentially, we can see that most of the methods use 2D segmentation on slice-by-slice, then they are interpolated to get the 3D volume. The interpolation plays a major role to avoid any loss of liver volume information. Sometimes, the concave shape of the contour being present on one slice is absent on the other and that could lead to a pseudo segmentation.
A segmentation approach usually considers the below three factors: (1) it should be user-friendly, (2) it should be accurate, and (3) it should be fast. Considering the pros and cons of the methods, region growing methods fulfill the segmentation criteria to a larger extent. This can be made more effective if it is combined with methods such as knowledge-based. Several published articles report such possible combinations. Then, the natural question arises: “why there has been not a single segmentation algorithm that could be fit for all?” Deep learning (DL) has garnered immense popularity since some years owing to its salient advantages, such as high accuracy and automatic nature of the network resulting the less involvement of the user (Ansari et al. 2022a, 2023). Specifically, the DL are getting attention, because the medical data is abundantly available these days. However, the DL methods have still not found that space to replace the conventional methods. This is because of a few key reasons: (1) labelled data are necessary for a robust DL method and a substantial amount of effort is needed to label the huge medical data, (2) high-power computational machine is needed to build and run the DL model; unfortunately, such high-performance work stations are rarely available in hospital set-up, (3) a DL model trained on the data available from a machine (say, Siemens) might not work as expected on a dataset obtained from a different machine (say, Phillips), this problem is known as domain generalization.
5.3 Implications and research challenges
Although there are many image segmentation algorithms, it is not clear which one is the best. We have already described the several advantages of conventional methods. Hopefully, this survey could help the readers to get the holistic view of all the methods and the suitability to be used. The accuracy of these methods has remained a concern for sure; however, if this could well be taken care of, then these conventional methods could truly be effective considering their low computational requirements. An inaccurate segmentation method could be the cause of lesion relapse in hepatic patients (Akhtar et al. 2021), thus, it is certainly not desirable. Further, we suggest for a personalized image segmentation framework, where the conventional segmentation method blocks could be dragged and dropped into the framework or pipeline. If this can possibly be accomplished, these conventional methods could truly be popular since the clinicians usually consider the AI methods as black box as compared to conventional methods. However, the input data, knowledge of existing algorithms, and their performances remain as crucial for developing new robust algorithms. Most of the authors test their algorithms on private data, thus, a fair comparison with the earlier published methods is certainly difficult. Unfortunately, the researchers do give little attention to this problem and keep focusing on developing new methods. Sometimes, the researchers struggle to compare their algorithms because of the lack of information about the workstation used. For instance, two similar algorithms performed on two different workstations (one high- and one low-power) would provide different performances, especially with respect to time. Considering the above challenges, if the research community is encouraged to follow some standard guidelines to post their databases used in their research paper online, then the whole community could access and get the benefits, especially, a new research methodology can easily be compared helping the researchers to continually improving the quality. The last but not the least, we have observed that a few methods forget to publish all the information, thus, these papers are hard to follow although the idea seems to be interesting. Thus, a researcher has to recode it again and obtain the results. Therefore, it may be recommended to encourage the researchers to make their data and codes public. This is necessary to save the time and effort of the researchers to not reinvent the wheel and the researchers could probably think onwards.
5.4 Limitations of the study
There have been some limitations in this study, especially with respect to the number of studies. Since we have targeted a wide spectrum of research papers, from the year 2000 to 2022, we must have missed several papers. It was quite difficult for us to include every journal that publishes on liver segmentation; we had to follow a plain protocol and include those articles that come under this protocol. The page constraint was another reason to discard these papers. We could have probably included a few more clinicians in the study to add more values to the survey by providing diverse clinical inputs. There are some other pressing issues as well from clinical standpoint, for instance, the explainability aspect, that we could not consider in this review. This issue has received significant attention recently after the AI models drew significant attentions, but we believe that the issue has always been present, in conventional methods too. This terminology was rarely highlighted earlier, thus, this is usually not found in conventional segmentation articles.
6 Conclusion
This study has presented a survey on an important computer aided diagnosis tool, segmentation. First, we have discussed the clinical relevance of image segmentation method. Then we have discussed the conventional automatic and semi-automatic approaches used over the past 20 years to segment liver images from CT scans. The methods have been critically evaluated from the view of their practical utility. We have summarized the segmentation evaluation metrics and it is found that an automatic method are better and but not desirable by the clinicians because they can rarely modify an automatic method, because they are end-to-end automatic. From that sense, a semi-automatic technique appears preferable since it allows the user (a clinician in this case) to direct the resultant shape to produce the desired outcome, but they are less accurate. Despite years of experimentation with various segmentation methods, various other issues still remain that we have listed in this paper. We have suggested a framework, where an effort could be made to build a personalized segmentation framework or customize a segmentation framework utilizing the conventional segmentation methods in an immanently free environment, which could probably reduce these tradeoffs between automatic and semiautomatic methods. Finally, we have included some suggestion: the researchers should probably be encouraged to make their data and code public.This could potentially help the computer vision community to be more focused addressing the genuine research problems rather than putting efforts on something that is probably unnecessary if the previous codes are accessible. If this happens, the researchers would probably be able to only look onwards.
Availability of data and materials
As no data sets were created or examined during the present investigation, data sharing is not relevant to this paper.
References
Akhtar Y, Dakua SP, Abdalla A, Aboumarzouk OM, Ansari MY, Abinahed J, Elakkad MSM, Al-Ansari A (2021) Risk assessment of computer-aided diagnostic software for hepatic resection. IEEE Trans Radiat Plasma Med Sci. https://doi.org/10.1109/TRPMS.2021.3071148
Al-Kababji A, Bensaali F, Dakua SP (2022) Scheduling techniques for liver segmentation: reducelronplateau vs. onecyclelr. In: Bennour A, Ensari T, Kessentini Y, Eom S (eds) Intelligent systems and pattern recognition. Springer, Cham, pp 204–212
Al-Kababji A, Bensaali F, Dakua SP, Himeur Y (2023) Automated liver tissues delineation techniques: a systematic survey on machine learning current trends and future orientations. Eng Appl Artif Intell. https://doi.org/10.1016/j.engappai.2022.105532
Amartur SC, Piraino D, Takefuji Y (1992) Optimization neural networks for the segmentation of magnetic resonance images. IEEE Trans Med Imaging 11:215–220
Ansari MY, Yang Y, Balakrishnan S, Abinahed J, Al-Ansari A, Warfa M, Almokdad O, Barah A, Omer A, Singh AV, Meher PK, Bhadra J, Halabi O, Azampour MF, Navab N, Wendler T, Dakua SP (2022a) A lightweight neural network with multiscale feature enhancement for liver CT segmentation. Sci Rep 12(1):14153. https://doi.org/10.1038/s41598-022-16828-6
Ansari MY, Abdalla A, Ansari MY, Ansari MI, Malluhi B, Mohanty S, Mishra S, Singh SS, Abinahed J, Al-Ansari A, Balakrishnan S, Dakua SP (2022b) Practical utility of liver segmentation methods in clinical surgeries and interventions. BMC Med Imaging 22(1):97. https://doi.org/10.1186/s12880-022-00825-2
Ansari MY, Yang Y, Meher PK, Dakua SP (2023) Dense-PSP-UNET: a neural network for fast inference liver ultrasound segmentation. Comput Biol Med 153:106478. https://doi.org/10.1016/j.compbiomed.2022.106478
Arora A, Seth S, Sharma Acharya S, Mukhopadhayaya S (1991) Case report: unusual CT appearances in a case of Budd–Chiari syndrome. Acad Radiol 43:431–432
Badakhshannoory H, Saeedi P (2011) A model-based validation scheme for organ segmentation in CT scan volumes. IEEE Trans Biomed Eng 58(9):2681–2693. https://doi.org/10.1109/TBME.2011.2161987
Beichel G, Gotschuli R, Sorantin E (2002) Diaphragm dome surface segmentation in CT data sets: a 3D active appearance model approach. Prog Biomed Opt Imaging 3:475–484
Besag J (1986) On the statistical analysis of dirty pictures. J R Stat Soc B 48:259–302
Bezdek JC, Hall LO, Clarke L (1993) Review of MR image segmentation techniques using pattern recognition. Med Phys 20:1033–1048
Bookstein F (1989) Principal warps: thin-plate splines and the decomposition of deformations. IEEE Trans Pattern Anal Mach Intell 6:567–585
Boykov Y, Kolmogorov V (2004) An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans Pattern Anal Mach Intell 26:1124–1137
Brandt ME, Bohant TP, Kramer LA, Fletcher JM (1994) Estimation of CSF, white and gray matter volumes in hydrocephalic children using fuzzy clustering of MR images. Comput Med Imaging Graph 18:25–34
Brown L (1994) A survey of image registration techniques. ACM Comput Surv 4:325–376
Campadelli CEP, Esposito A (2009) Liver segmentation from computed tomography scans: a survey and a new algorithm. Artif Intell Med 45:185–196
Casciaro S, Franchini R, Massoptier L, Casciaro E, Conversano F, Malvasi A, Lay-Ekuakille A (2011) Fully automatic segmentations of liver and hepatic tumors from 3-D computed tomography abdominal images: comparative evaluation of two automatic methods. IEEE Sens J PP(99):1. https://doi.org/10.1109/JSEN.2011.2108281
Casciaro S, Franchini R, Massoptier L, Casciaro E, Conversano F, Malvasi A, Lay-Ekuakille A (2012) Fully automatic segmentations of liver and hepatic tumors from 3-D computed tomography abdominal images: comparative evaluation of two automatic methods. IEEE Sens J 12(3):464–473. https://doi.org/10.1109/JSEN.2011.2108281
Caselles KRV, Sapiro G (1997) Geodesic active contours. Int J Comut Vis 22:61–79
Chartrand G, Cresson T, Chav R, Gotra A, Tang A, De Guise JA (2016) Liver segmentation on CT and MR using Laplacian mesh optimization. IEEE Trans Biomed Eng 64(9):2110–2121
Chen X, Udupa JK, Bagci U, Zhuge Y, Yao J (2012) Medical image segmentation by combining graph cuts and oriented active appearance models. IEEE Trans Image Process 21(4):2035–2046
Choi D, Haynor H, Kim Y (1991) Partial volume tissue classification of multichannel magnetic resonance images—a mixed model. IEEE Trans Med Imaging 10:395–407
Choudhary A, Moretto N, Ferrarese F, Zamboni G (2008) An entropy based multi-thresholding method for semi-automatic segmentation of liver tumors. In: 3D segmentation in the clinic: a grand challenge
Clarke LP, Velthuizen RP, Camacho MA, Heine JJ, Vaidyanathan M, Hall LO, Thatcher RW, Silbiger ML (1995) MRI segmentation: methods and applications. Magn Reson Imaging 13:343–368
Dakua SP (2013a) Performance divergence with data discrepancy: a review. Artif Intell Rev 40:429–455. https://doi.org/10.1007/s10462-011-9289-8
Dakua SP (2013b) Use of chaos concept in medical image segmentation. Comput Methods Biomech Biomed Eng Imaging Visual 1(1):28–36. https://doi.org/10.1080/21681163.2013.765709
Dakua SP (2014) Annularcut: a graph-cut design for left ventricle segmentation from magnetic resonance images. IET Image Proc 8:1–1110
Dakua SP (2017) Towards left ventricle segmentation from magnetic resonance images. IEEE Sens J 17(18):5971–5981. https://doi.org/10.1109/JSEN.2017.2736641
Dakua SP, Abi-Nahed J (2013) Patient oriented graph-based image segmentation. Biomed Signal Process Control 8(3):325–332. https://doi.org/10.1016/j.bspc.2012.11.009
Dakua SP, Nayak A (2022) A review on treatments of hepatocellular carcinoma—role of radio wave ablation and possible improvements. Egypt Liver J 12(1):30. https://doi.org/10.1186/s43066-022-00191-2
Dakua SP, Sahambi JS (2009) LV contour extraction from cardiac MR images using random walks approach. In: 2009 IEEE international advance computing conference, pp 228–233. https://doi.org/10.1109/IADCC.2009.4809012
Dakua SP, Abi-Nahed J, Al-ansari A (2018) A PCA-based approach for brain aneurysm segmentation. Multidim Syst Sign Process 29(4):257–277
Fan S, Lo C, Liu C, Yong B, Chan J, Ng L (2000) Safety of donors in live donor liver transplantation using right lobe grafts. Arch Surg 135:336–340
Fogarollo S, Bale R, Harders M (2023) Towards liver segmentation in the wild via contrastive distillation. Int J Comput Assist Radiol Surg 18(7):1143–1149. https://doi.org/10.1007/s11548-023-02912-3
Foruzan A, Zoroofi R, Hori M, Sato Y (2009a) Liver segmentation by intensity analysis and anatomical information in multi-slice CT images. Int J Comput Assist Radiol Surg 4:287–297
Foruzana A, Zoroofia R, Horib M, Satoc Y (2009b) A knowledge-based technique for liver segmentation in CT data. Comput Med Imaging Graph 33(33):567–587
Gao KAJ, Kak A (2005) A deformable model for automatic CT liver extraction. Acad Radiol 12:1178–1189
Goryawala M, Guillen MR, Cabrerizo M, Barreto A, Gulec S, Barot TC, Suthar RR, Bhatt RN, Mcgoron A, Adjouadi M (2011) A 3-D liver segmentation method with parallel computing for selective internal radiation therapy. IEEE Trans Inf Technol Biomed 16(1):62–69
Halabi O, Balakrishnan S, Dakua SP, Navab N, Warfa M (2020) Virtual and augmented reality in surgery. In: Doorsamy W, Paul BS, Marwala T (eds) Virtual and augmented reality in surgery. Springer, Cham, pp 257–285. https://doi.org/10.1007/978-3-030-48230-5_11
Hall LO, Bensaid AM, Clarke LP, Velthuizen RP, Silbiger MS, Bezdek JC (1992) A comparison of neural network and fuzzy clustering techniques in segmenting magnetic resonance images of the brain. IEEE Trans Neural Netw 3:672–682
Harris GJ, Rhew EH, Noga T, Pearlson GD (1991) User-friendly method for rapid brain and CSF volume calculation using transaxial MRI images. Psychiatry Res Neuroimaging 40:61–68
Heimann T, Van Ginneken B, Styner MA, Arzhaeva Y, Aurich V, Bauer C, Beck A, Becker C, Beichel R, Bekes G, Bello F (2009) Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging 28:1251–1265
Hsu LCC, Chen C (2008) Automatic segmentation of liver pet images. Comput Med Imaging Graph 32:610
Ji H, He J, Yang X, Deklerck R, Cornelis J (2013) ACM-based automatic liver segmentation from 3-D CT images by combining multiple atlases and improved mean-shift techniques. IEEE J Biomed Health Inf 17(3):690–698
Jin R, Wang M, Xu L, Lu J, Song E, Ma G (2023) Automatic 3D CT liver segmentation based on fast global minimization of probabilistic active contour. Med Phys 50(4):2100–2120. https://doi.org/10.1002/mp.16116
Kohn MI, Tanna NK, Herman GT, Resnick SM, Mozley PD, Gur RE, Alavi A, Zimmerman RA, Gur RC (1991) Analysis of brain and cerebrospinal fluid volumes with MR image part 1: methods, reliability, and validation. Radiology 178:115–122
Kolmogorov V, Zabih R (2004) What energy functions can be minimized via graph cuts? IEEE Trans Pattern Anal Mach Intell 26:147–159
Le DC, Chinnasarn K, Chansangrat J, Keeratibharat N, Horkaew P (2021) Semi-automatic liver segmentation based on probabilistic models and anatomical constraints. Sci Rep 11(1):1–19
Lee W, Chen Y, Chen Y, Hseih K (2005) Unsupervised segmentation of ultrasonic liver images by multi resolution fractal feature vector. Inf Sci 175:177–199
Li C, Lee C (1993) Minimum cross entropy thresholding. Pattern Recogn 26:617–625
Li C, Wang X, Li J, Eberl S, Fulham M, Yin Y, Feng DD (2012) Joint probabilistic model of shape and intensity for multiple abdominal organ segmentation from volumetric CT images. IEEE J Biomed Health Inf 17(1):92–102
Li C, Wang X, Eberl S, Fulham M, Yin Y, Chen J, Feng DD (2013) A likelihood and local constraint level set model for liver tumor segmentation from CT volumes. IEEE Trans Biomed Eng 60(10):2967–2977
Li G, Chen X, Shi F, Zhu W, Tian J, Xiang D (2015) Automatic liver segmentation based on shape constraints and deformable graph cut in CT images. IEEE Trans Image Process 24(12):5315–5329
Liang Z (1993) Tissue classification and segmentation of MR images. IEEE Eng Med Biol Mag 1993:81–85
Liang Z, Jaszczak R, Coleman R (1992) Parameter estimation of finite mixtures using the EM algorithm and information criteria with application to medical image processing. IEEE Trans Nucl Sci 39:1126–1133
Liang Z, MacFall J, Harrington D (1994) Parameter estimation and tissue segmentation from multispectral MR images. IEEE Trans Med Imaging 13:339–348
Lim JYS, Ho Y (2005) Segmentation of the liver using the deformable contour method on CT images. LNCS 3767:570–581
Lim K, Pfefferbaum A (1989) Segmentation of MR brain images into cerebrospinal fluid spaces, white and gray matter. J Comput Assist Tomogr 13:588–593
Lin WC, Tsao EW, Chen C (1992) Constraint satisfaction neural networks for image segmentation. Pattern Recogn 25:679–693
Linguraru MG, Sandberg JK, Li Z, Pura JA, Summers RM (2009) Atlas-based automated segmentation of spleen and liver using adaptive enhancement estimation. In: MICCAI 2009, Part II, LNCS, vol 5762, pp 1001–1008
Linguraru M, Sandberg J, Li Z, Shah F, Summers R (2010) Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation. Med Phys 37:771–783
Liu F, Zhao B, Kijewski PK, Wang L, Schwartz LH (2005) Liver segmentation for CT images using GVF snake. Med Phys 32:3699
Lundervold A, Storvik S (1995) Segmentation of brain parenchyma and cerebrospinal fluid in multispectral magnetic resonance images. IEEE Trans Med Imaging 14:339–348
Massoptier L, Casciaro S (2008) A new fully automatic and robust algorithm for fast segmentation of liver tissue and tumours from CT scans. Eur Radiol 18:1658–1665
Mohanty S, Dakua SP (2022) Toward computing cross-modality symmetric non-rigid medical image registration. IEEE Access 10:24528–24539. https://doi.org/10.1109/ACCESS.2022.3154771
Montagnat J, Delingette H (1996) Volumetric medical images segmentation using shape constrained deformable models. In: CVRMed-MRCAS. Springer, Berlin, pp 13–22
Morrison M, Attikiouzel Y (1994) An introduction to the segmentation of magnetic resonance medical images. Aust Comput J 26:90–98
Nakayama Y, Li Q, Katsuragawa S, Ikeda R, Hiai Y, Awai K et al (2006) Automated hepatic volumetry for living related liver transplantation at multisection CT. Radiology 240:743–748
Okada Yokota T, Hori K, Nakamoto M, Nakamura MH, Sato Y (2008) Construction of hierarchical multi-organ statistical atlases and their application to multiorgan segmentation from CT images. In: MICCAI 2008, part I. LNCS, vol 5241. Springer, Berlin
Ortendahl DA, Crooke LE, Hylton NM, Kaufman L (1985) Tissue characterization using intrinsic NMR parameters and a hierarchical processing algorithm. IEEE Trans Nucl Sci NS–32:875–879
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9:62–66
Ozkan M, Dawant BM, Maciunas RJ (1993) Neural-network based segmentation of multi-modal medical images: a comparative and prospective study. IEEE Trans Med Imaging 12:534–544
Park H, Bland PH, Meyer CR (2003) Construction of an abdominal probabilistic atlas and its application in segmentation. IEEE Trans Med Imaging 22(4):483–492. https://doi.org/10.1109/TMI.2003.809139
Perona P, Malik J (1990) Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell 12:629–639
Pham D, Xu C, Prince J (2000) Current methods in medical image segmentation. Annu Rev Biomed Eng 2:315–337
Rai P, Dakua S, Abinahed J, Balakrishnan S (2021) Feasibility and efficacy of fusion imaging systems for immediate post ablation assessment of liver neoplasms: protocol for a rapid systematic review. Int J Surg Protocols 25(1):209–215. https://doi.org/10.29337/ijsp.162
Rueckert D et al (1999) Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging 18:712–721
Rusko L, Bekes G, Fidrich M (2009) Automatic segmentation of the liver from multi- and single-phase contrast-enhanced CT images. Med Image Anal 13(6):871–882. https://doi.org/10.1016/j.media.2009.07.009 (Includes special section on computational biomechanics for medicine)
Santago P, Gage H (1993) Quantification of MR brain images by mixture density and partial volume modeling. IEEE Trans Med Imaging 12:566–574
Schmidt M, Athelogou G, Schonmeyer M, Korn RR, Binnig G (2007) Cognition network technology for a fully automated 3D segmentation of liver. In: Workshop in 3D segmentation in the clinic: a grand challenge, vol 1. MICCAI, pp 125–134
Schmidt G, Binnig G, Kietzmann M, Kim J (2008) Cognition network technology for a fully automated 3D segmentation of liver tumors. In: 3D segmentation in the clinic: a grand challenge. MICCAI
Selver M, Kocaoglu A, Demir G, Dogan H, Dicle O, Güzelis C (2008) Patient oriented and robust automatic liver segmentation for pre-evaluation of liver transplantation. Comput Biol Med 38:765–784
Sethian J (1996) A fast marching level set method for monotonically advancing fronts. Proc Natl Acad Sci USA 93:1591–1595
Singh AV, Chandrasekar V, Paudel N, Laux P, Luch A, Gemmati D, Tisato V, Prabhu KS, Uddin S, Dakua SP (2023) Integrative toxicogenomics: advancing precision medicine and toxicology through artificial intelligence and omics technology. Biomed Pharmacother 163:114784. https://doi.org/10.1016/j.biopha.2023.114784
Soler L, Delingette H, Malandain G, Montagnat J, Ayache N, Koehl C, Dourthe O, Malassagne B, Smith M, Mutter D et al (2001) Fully automatic anatomical, pathological, and functional segmentation from CT scans for hepatic surgery. Comput Aided Surg 6(3):131–142
Song X, Deng G, Zhuang Y, Zeng N (2019) An improved confidence connected liver segmentation method based on three views of CT images. IEEE Access 7:58429–58434
Studholme HDC, Hawkes D (1999) An overlap invariant entropy measure of 3D medical image alignment. Pattern Recogn 32:71–86
Suzuki K, Kohlbrenner R, Epstein M, Obajuluwa A, Xu J, Hori M (2010) Computer-aided measurement of liver volumes in CT by means of geodesic active contour segmentation coupled with level-set algorithms. Med Phys 37:2159–2166
Taxt T, Lundervold A (1994) Multispectral analysis of the brain using magnetic resonance imaging. IEEE Trans Med Imaging 13:470–481
Vincent L, Soille P (1991) Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans Pattern Anal Mach Intell 13:583–598
Wang S, Fu D, Xu M, Hu D (2007) Advanced fuzzy cellular neural network: application to CT liver images. Artif Intell Med 39(1):65–77
Whiteley M, Welsh B, Roggemann M (1998) Limitations of gaussian assumptions for the irradiance distribution in digital imagery: nonstationary image ensemble considerations. J Opt Soc Am A 15:802–810
Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D (2013) Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans Med Imaging 32(9):1723–1730
Yuan F, Chi Y, Huang S, Liu J (2011) Modeling \(n\)-furcated liver vessels from a 3-D segmented volume using hole-making and subdivision methods. IEEE Trans Biomed Eng 59(2):552–561
Zhai X, Eslami M, Hussein ES, Filali MS, Shalaby ST, Amira A, Bensaali F, Dakua S, Abinahed J, Al-Ansari A, Ahmed AZ (2018) Real-time automated image segmentation technique for cerebral aneurysm on reconfigurable system-on-chip. J Comput Sci 27:35–45. https://doi.org/10.1016/j.jocs.2018.05.002
Zhai X, Amira A, Bensaali F, Al-Shibani A, Al-Nassr A, El-Sayed A, Eslami M, Dakua SP, Abinahed J (2019) ZYNQ SOC based acceleration of the lattice Boltzmann method. Concurr Comput Pract Exp 31(17):5184. https://doi.org/10.1002/cpe.5184
Zhang X, Tian J, Deng K, Wu Y, Li X (2010) Automatic liver segmentation using a statistical shape model with optimal surface detection. IEEE Trans Biomed Eng 57(10):2622–2626. https://doi.org/10.1109/TBME.2010.2056369
Zhang Q, Fan Y, Wan J, Liu Y (2017) An efficient and clinical-oriented 3D liver segmentation method. IEEE Access 5:18737–18744
Zhou X, Kitagawa T, Okuo K, Hara T, Fujita H, Yokoyama R, Kanematsu M, Hoshi H (2005) Construction of a probabilistic atlas for automated liver segmentation in non-contrast torso CT images. Int Cong Ser 1281:1169–1174. https://doi.org/10.1016/j.ics.2005.03.079 (CARS 2005: computer assisted radiology and surgery)
Acknowledgements
This work was supported in part by the Qatar National Research Fund (a member of Qatar Foundation) under Grant NPRP-11 S-1219-170106; in part by the Medical Research Center, Hamad Medical Corporation Doha, Qatar, under Grant MRC-01-19-327. The authors alone bear full responsibility for the findings, which are a reflection of their work.
Funding
Open Access funding provided by the Qatar National Library. This work was supported in part by the Qatar National Research Fund (a member of Qatar Foundation) under Grant NPRP-11S-1219-170106; in part by the Medical Research Center, Hamad Medical Corporation Doha, Qatar, under Grant MRC-01-19-327. The authors alone bear full responsibility for the findings, which are a reflection of their work. The Open Access funding was provided by the Qatar National Library.
Author information
Authors and Affiliations
Contributions
SPD, LJ, SIA, and GKM have conceptualized the study and participated in design of the study, and manuscript review, while RKM, DCL, and AMIA have participated in data analysis, design of the study, manuscript preparation, literature search, and manuscript review.
Corresponding author
Ethics declarations
Conflict of interest
The authors suggest no conflict of interest. The study’s design, data collection, analysis, or interpretation; the preparation of the paper; or the choice to publish the findings were all made independently of the funders.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethics approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohapatra, R.K., Jolly, L., Lyngdoh, D.C. et al. A comprehensive survey to study the utilities of image segmentation methods in clinical routine. Netw Model Anal Health Inform Bioinforma 13, 2 (2024). https://doi.org/10.1007/s13721-023-00436-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-023-00436-z