Abstract
Sex determination is one of the major developmental events in higher metazoans, where complex molecular mechanisms define two physiologically and behaviorally distinct organisms still genetically compatible. In honey bees, the sex determination cascade is initiated by the allelic composition of complementary sex determiner (csd) gene: males develop from hemi or homozygous embryos, whereas females develop from heterozygous embryos. In females, different alleles of csd lead to the formation of female-specific variants of feminizer (fem) and doublesex (dsx). In males, male-specific variants of fem and dsx are formed by default. In this paper, we investigated the genes of sex determination in the stingless bees Melipona quadrifasciata, Scaptotrigona postica, and Frieseomelitta varia. Our results revealed that the architecture of fem and dsx transcripts is highly conserved among the three stingless bee species, and also with honey bees.
Similar content being viewed by others
1 Introduction
The sex determination system in the eusocial Hymenoptera is based on the single locus complementary sex determination (Woyke 1965; de Camargo 1979; Kerr 1987; Paxton et al. 2003), in which diploid and heterozygotic eggs give rise to females while haploid (hemizygotic) or diploid homozygotic eggs give rise to males (Dzierzon 1845). The complexity of the molecular events underlying the molecular mechanisms of sex determination was first hypothesized by Kerr (1974). He combined the genetic basis of honey bee sex determination with the Britten and Davidson (1969) theory and proposed two hypotheses: (1) the sex alleles are complementary and produce a functional activator RNA if the alleles are different and (2) the functional activator RNA acts on the production of polypeptides essential to trigger ovary development. The idea of complementary sex alleles was later confirmed in honey bees after the identification of 19 sex alleles (Adams et al. 1977) and functional assays showed that the complementary sex determiner gene was the primary signal for sex determination pathway (Beye et al. 2003; Beye 2004). In heterozygotic embryos for the csd locus, heterodimer of Csd directs the female-specific splicing of feminizer (fem) transcripts and produces the active protein Fem (FemF). In turn, FemF controls the splicing of the doublesex (dsx) transcripts which leads to the formation of female-specific transcripts (dsxF) that controls female differentiation (Cristino et al. 2006; Hasselmann et al. 2008; Gempe et al. 2009). In homo- or hemizygotic embryos, the absence of a heterodimeric form of Csd protein results in the formation of a male-specific transcript of the gene fem (femM), which produces a truncated and non-functional protein. Thus, by default, male-specific transcripts of dsx are formed and DsxM directs male differentiation.
Consistent with the “bottom-up” hypothesis of sex determination pathways (Wilkins 1995), csd is exclusively found in Apis sp. and arose by duplication of fem (Hasselmann et al. 2008), while fem and dsx have been found in all bees with sequenced genomes (Kapheim et al. 2015). The gene dsx is highly conserved within insects, vertebrates, and nematodes and encodes a zinc-finger transcription factor (DSX) that presents the DM domain and a dimer domain (Burtis 2002). Both csd and fem code for proteins rich in serine and arginine (SR-related proteins) (Beye et al. 2003; Beye 2004; Hasselmann et al. 2008), which are known to participate in RNA splicing through the recognition of regulatory elements in exons of primary messenger RNAs (Shepard and Hertel 2009). While the exact mechanism by which fem controls dsx splicing in honey bee is still unknown, in Drosophila, the protein coded by transformer (a homolog of fem) binds to regulatory elements in the female-specific exon of dsx and signals to the splicing machinery to include the female exon in the mature mRNA (Hedley and Maniatis 1991; Heinrichs et al. 1998).
European honey bees are the best studied Hymenoptera but are far from representative of the diversity of this group. Stingless bees (Meliponini) display a rich diversity of life stories and social organization and differ from honey bees in several aspects. The most striking differences are the mating system, caste determination, and participation of workers in the production of males. Honey bee queens mate multiple times (Estoup et al. 1994; Oldroyd et al. 1997) in contrast to the single-mated stingless bee queens (Peters et al. 1999). Caste determination in honey bees is triggered by quality of food while in stingless bees, caste determination is based on mass-provisioning (for example, Frieseomelitta varia and Scaptotrigona postica) and can be affected by a genetic component (Melipona quadrifasciata) (Hartfelder et al. 2006). Honey bee workers rarely lay eggs and stingless bee workers display diverse strategies that ranges from never laying egg (Frieseomelitta varia) (Boleli et al. 1999) to being responsible for 20 to 60% of male production (Scaptotrigona postica and Melipona quadrifasciata, respectively) (Tóth et al. 2002). This richness in diversity of stingless bees’ biology prompted us to investigate the sex determination cascade in three species of stingless bees which have major differences in their social organization.
To provide further information on the sex determination pathways of stingless bees, we characterized the architecture of sex-specific transcripts of fem and dsx in males and females of F. varia, M. quadrifasciata, and S. postica. The combination of exons to form both female and male transcripts is highly conserved among the stingless and honey bees. Additionally, the observation of conserved short repeats in the female-specific exon of dsx in stingless bees and honey bees is consistent with recognition sites for SR-rich proteins (like Fem) and supports the hypothesis of conservation of the mechanisms that regulate the alternative splicing of sex determination genes in bees.
2 Methods
2.1 Sampling of stingless bees
Adults of workers and males of the stingless bees F. varia and S. postica were sampled directly from colonies kept in the meliponary at the University of São Paulo, Ribeirão Preto, Brazil. Newly emerged adults of M. quadrifasciata were sampled from colonies kept in the meliponary at the University of São Paulo, São Paulo, Brazil. Each sex for each specie was represented in triplicates, individually stored in Trizol ® (Invitrogen) and kept at − 80 °C until RNA extraction.
2.2 Extraction of total RNA and amplification of sex-specific transcripts
Total RNA was purified using TRIZOL® (Invitrogen) according to producer’s protocol. First-strand cDNA was synthesized by reverse transcription from 3 μg of total RNA using 100 units of Super Script II reverse transcriptase (Thermo Fisher) and 2.0 pmol/μL oligo (dT12-18) primer (Thermo Fisher). Primers used to amplify fem and dsx transcripts of the three selected stingless bee species were generated based on M. quadrifasciata sequences using Primer 3 tool (Rozen and Skaletsky 2000) (Table I and Table S1). PCR reactions were performed using 10 pmol/μL of forward and reverse primers, 1.0 μL of first-strand cDNA (diluted 1:5 or 1:10), Master mix (Promega), or Platinum® Taq DNA Polymerase High Fidelity (Thermo Fisher). The amplification products were analyzed by electrophoresis on 1.5% agarose gels containing UniSafe Dye (Uniscience).
2.3 Cloning and DNA sequencing
Amplicons of fem and dsx from F. varia, M. quadrifasciata, and S. postica were purified from agarose gel and cloned using pGEM-T easy plasmid kit (Promega). Three to five insert-containing plasmids of each transcript (for each species) were subjected to the dideoxy sequencing method and ABI 3500xL Genetic Analyzer (Applied Biosystems) using the M13-reverse and M13-forward universal primers. The quality of the fragments and the assembly was performed by Geneouis 11.0.4 (http://ww.geneious.com, Kearse et al. (2012)).
2.4 Computational analysis of sex-specific transcripts of fem and dsx genes
To obtain the complete architecture of the sex-specific transcripts, the sequences of the amplified fragments were merged with the predicted gene models of fem and dsx for F. varia and M. quadrifasciata. In general, alternative transcripts are not predicted in first drafts of genomes. Thus, the predicted sequences for feminizer and doublesex genes (gene models) in M. quadrifasciata and F. varia do not include alternative usage of exons. Using cDNA samples of males and females to amplify fragments of feminizer and doublesex transcripts, we were able to characterize the sex-specific exon usage in the three analyzed species. The amplified fragments did not include the entire extension of the genes; thus, to obtain full-length transcripts, we merged the gene model sequences (predicted annotation of the genes feminizer and doublesex) with the sex-specific fragments we have amplified and sequenced. Transcript architectures were inferred by aligning the sequenced fragments against M. quadrifasciata and F. varia genomes using the BLAST tool available at Hymenoptera Genome Database. Transcripts of S. postica were inferred by alignment against M. quadrifasciata genome; thus, the length of intronic regions could not be inferred. BLAST results were plotted using Ugene platform (Okonechnikov et al. 2012). Protein sequences were inferred using the online tool EMBOSS-Transeq (Li et al. 2015). Sequences of fem and dsx transcripts obtained for F. varia, M. quadrifasciata, and S. postica were deposited into the GenBank database (Clark et al. 2016) (Table S2). The identification of conserved domains was performed using NCBI’s Conserved Domain Database (Marchler-Bauer et al. 2014). The percentage of similarity between domains of different bee species was calculated using the online tool Sequence Identity And Similarity (http://imed.med.ucm.es/Tools/sias.html).
2.5 Search for regulatory elements for SR-related proteins in female-specific exon of dsx
To search for putative regulatory elements for SR-related proteins, we aligned the female-specific exon of dsx transcripts of A. mellifera, M. quadrifasciata, F. varia, and S. postica using MEGA v. 7 (Kumar et al. 2016). Then, we actively searched for the regulatory elements found in the fifth exon of honey bee dsx (G/U)GAAGAU(A/U) (Bertossa et al. 2009) in the female-specific exon of dsx in stingless bees. The regulatory elements we found were represented using a logo plotted by the online tool WebLogo 3 (http://weblogo.threeplusone.com/create.cgi).
3 Results
3.1 Sex-specific transcripts of feminizer and doublesex in stingless bees
Expression of sex-specific transcripts for both feminizer and doublesex genes were observed from cDNA samples of F. varia, M. quadrifasciata, and S. postica (Figure 1). To uncover the architecture of these sex-specific transcripts in stingless bees, we sequenced and mapped the transcripts of both feminizer and doublesex genes of F. varia, M. quadrifasciata, and S. postica. The architecture of male- and female-specific transcripts of both fem and dsx genes was highly consistent among F. varia, M. quadrifasciata, and S. postica (Figures 2 and 3). In both F. varia and M. quadrifasciata, female-specific transcripts of fem are composed by eight exons and male-specific transcripts are composed of nine exons (Figure 2a). The inclusion of exon 3 in the male-specific transcripts of fem inserts a premature stop codon and produces a truncated Fem protein in males of both species (Figure 2a). Similar architecture was observed for S. postica as the partial sequences of fem transcripts revealed that the inclusion of an extra exon in male-specific transcripts inserts a premature stop codon in the coding sequence. A Sex Determination Protein N terminal domain (SDP N) was identified in the first exon of both female- and male-specific transcripts and a SR-rich domain was found in exon 6 of female-transcripts of F. varia and M. quadrifasciata (Figure 2b). The protein sequence inferred from both female- and male-specific transcripts of fem gene has similar length among stingless bees and honey bees (Table II). The SDP domain shared by both female- and male-specific proteins is slightly longer in stingless bee species (161 amino acids in M. quadrifasciata and 163 amino acids in F. varia) compared to honey bees (153 amino acids in A. mellifera) (Figure S1). The comparison of SDP N domain sequence among different bee species revealed moderate similarity between A. mellifera and F. varia (47.33%, Figure S1) and lower similarity between A. mellifera and M. quadrifasciata (27.33%) and between M. quadrifasciata and F. varia (28.57%). The SR-rich region, specific of FemF, is also longer in stingless bees (58 and 52 amino acids long in M. quadrifasciata and F. varia, respectively) compared to honey bees (44 amino acids in A. mellifera) (Figure S1). The SR-rich region is similar between M. quadrifasciata and F. varia (55.31%) but it is less similar between stingless bee species and honey bee species (similarity between F. varia and A. mellifera and between M. quadrifasciata and A. mellifera is 29.54% and 31.81%, respectively).
Expression of feminizer and doublesex in male and females of stingless bees. a Expression of sex-specific fem transcripts in female and males of F. varia, M. quadrifasciata, and S. postica. b Expression of sex-specific dsx transcripts in female and males of F. varia, M. quadrifasciata, and S. postica. DNA Ladder: 100 bp DNA Ladder (Invitrogen).
Transcripts and protein variants of feminizer gene in stingless bees. a Architecture of female- and male-specific fem transcripts reveals that the similar architecture is shared by F. varia and M. quadrifasciata. The architecture of fem transcripts of S. postica was inferred based on the alignment in F. varia genome. Male-specific transcripts of F. varia, M. quadrifasciata, and S. postica present an extra exon that introduces a premature stop codon and generates a truncated FemM protein. Boxes represent exons. The light green boxes are protein-coding exons. The regions highlighted in orange and red represent the SPD and SR-rich domains, respectively. Blue boxes highlight the male-specific exon that contains the premature stop codon. Gray boxes represent the non-translated exons. Numbers indicate the length of exons and introns in base pairs (bp). The arrows represent both forward and reverse primers. b Effects of sex-specific splicing events on protein outcomes. The region highlighted in orange represents the SDP domain in both female- and male-specific Fem protein, while the region highlighted in red represents SR-rich domain in female-specific Fem protein.
Transcripts and protein variants of doublesex gene in stingless bees. a Architecture of female- and male-specific dsx transcripts reveals that a similar architecture is shared by F. varia and M. quadrifasciata. The architecture of dsx transcripts of S. postica was inferred based on the alignment in F. varia genome. Female-specific transcripts present two extra exons in F. varia, M. quadrifasciata, and S. postica. Boxes represent exons. The light green boxes are protein-coding exons. The portion of the first exon marked by diagonal stripes defines the region occupied by the DM domain. The region highlighted in orange represents partial dsx dimer domain disrupted by the inclusion of exons 4 and 5 in female-specific dsx transcripts. The regions highlighted in red represent dsx dimer domain in male-specific dsx transcript. Gray boxes represent the non-translated exons. Numbers indicate the length of exons and introns in base pairs (bp). The arrows represent both forward and reverse primers. b Effects of sex-specific splicing events on protein outcomes. The region highlighted by diagonal stripes defines the region occupied by the DM domain. The region highlighted in orange represents partial dsx dimer domain in female-specific dsx protein and the regions highlighted in red represent dsx dimer domain in male-specific dsx protein.
Male-specific transcripts of dsx are composed by four exons (exons 1, 2, 3, and 6) and female-specific transcripts are composed of six exons (exons 1, 2, 3, 4, 5, and 6). The protein sequence of both female and male transcripts of dsx genes was inferred from the mRNA sequences. Fem and Dsx proteins have similar length among stingless bees and honey bees (Table II). The DM domain, which characterizes the gene of this family, was found in the first exon of both male- and female-specific transcripts of dsx in each of the three species. The DM domain is 48 amino acids long in A. mellifera and 49 in both F. varia and M. quadrifasciata. It is identical between F. varia and M. quadrifasciata (100% similarity, Figure S2) but these diverge from DM domain of A. mellifera (14.58%, Figure S2). Note that the differences found between each of F. varia, M. quadrifasciata, and A. mellifera in comparison to S. postica are affected by the incompleteness of S. postica sequence. The complete dsx dimer domain of stingless bees DsxM protein is 56 amino acids long and is between 94.64 and 100% similar among themselves and with honey bees (Figure S2). The inclusion of exons 4 and 5 inserts a stop codon that disrupts the dsx dimer domain in DsxF for all analyzed species yielding a partial dsx dimer domain 42 amino acids long (Figure 3a, B). The partial dsx dimer domain of stingless bee DsxF is also highly similar between stingless bee species and honey bees (100 to 92.85%, Figure S2).
3.2 Female-specific exon of dsx transcripts may be regulated by SR-related protein
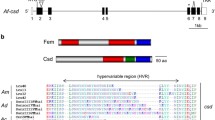
We searched for the short repeats (U/G)GAAGAU(U/A) in the female-specific exons of dsx transcripts in F. varia, M. quadrifasciata, and S. postica. Three short repeats (C/G)GAAGAU(C/U) were found in the 3′ end of the female-specific exon of F. varia and S. postica, and two were found in M. quadrifasciata (Figure 3a, b). The repeats found in the female-specific exon of dsx in the stingless bees are very similar to the repeats described for A. mellifera, which are UGAAGAU and GGAAGAA (Bertossa et al. 2009).
4 Discussion
In this paper, we investigated the genes of sex determination in the stingless bees F. varia, M. quadrifasciata, and S. postica, which have been poorly studied in comparison to honey bees. Stingless bees have a pantropical distribution and are important pollinators of native plants and crops (Viana et al. 2014). Stingless bees represent a valuable group to study because of their stunning diversity of biology life stories and reproductive behavior that includes colonies preferentially headed by one singly mated queen to colonies with transient episodes of multiple queens (as reported in Melipona bicolor, M. quadrifasciata, and F. varia) (Vollet-Neto et al. 2018). Here, we show that despite of the diversity in mating system and social organization, the regulation of sex determination cascade is conserved among stingless bee species and honey bees.
fem has been found in all bees with sequenced genomes (Kapheim et al. 2015), although its sex-specific transcripts have only been described for honey bees and Melipona interrupta (Hasselmann et al. 2008; Brito et al. 2015). In honey bees, functional studies showed that femF is crucial to the differentiation of ovaries, since its knockdown leads to the formation of testis in females (Gempe et al. 2009). Here, we show that transcripts of fem are sex-specific spliced and the architecture of female- and male-transcripts of fem gene is highly conserved between stingless bees and honey bees (HASSELMANN et al. 2008) (Figure 2a). Surprisingly, extra bands were amplified in female samples of F. varia (Figure 1a), indicating the potential formation of other alternative transcripts, which may be investigated in future studies. The analyses of the protein outcomes of both female- and male-specific transcripts revealed that FemF and FemM share the N-terminal SDP N domain and differ in the C-terminal region, resulting in the presence of a SR-rich region only in FemF (Figure 2b). The presence of both N-terminal SDP N domain and a C-terminal SR-rich region is characteristic of honey bee fem gene (Hasselmann et al. 2008) indicating conservation of the mechanism controlling sex-specific splicing events of fem transcripts among different bee species.
The most conserved gene at the bottom of the sex determination cascade, dsx, has been found in all insects investigated so far, including dipteran, hymenopteran, and lepidopterans, and also in the crustacean Daphnia magna (Kato et al. 2011). Here, we described sex-specific transcripts of dsx in F. varia, M. quadrifasciata, and S. postica. The architecture of the transcripts is highly conserved among the stingless bees (Figure 3a) and honey bees (Cristino et al. 2006; Cho et al. 2007). The female-specific dsx transcript is formed by the inclusion of one exon in honey bee and two exons in stingless bees that disrupt the dsx dimer domain, which results in a shorter protein compared to the protein coded by the male-specific transcript (Figure 3b). Stingless bees DsxF and DsxM proteins share an N-terminal DNA binding domain (DM domain) and the C-terminal domain, where the dsx dimer is located, differs (Figure 3b). A similar domain display was observed in DsxF and DsxM of honey bees (Cho et al. 2007) and Drosophila (Erdman et al. 1996). The DM domain is highly conserved among metazoan species (Raymond et al. 1998) and, in Drosophila, the recognition of DNA motifs by DM domain is enhanced by the C-terminal dimerization domain (dsx dimer domain) (Cho and Wensink 1998). The dsx dimer domain is conserved in insect species and is proposed to mediate recruitment of transcriptional co-regulatory factors (Erdman et al. 1996; Garrett-Engele et al. 2002). Upstream regions of genes bound by Drosophila DsxF and DsxM had mouse orthologs also bound by mouse Dsx pointing to a high degree of conservation of Dsx target across the animal kingdom (Clough et al. 2014). Although the specific targets of DsxF and DsxM are unknown for bees, it is likely that they also share common genes due to the conservation of both DM and dsx dimer domains.
The inclusion of the exclusive female-specific exon that disrupts the dsx dimer domain in stingless bees seems to require specific signalization to the splicing machinery mediated by SR-rich proteins as proposed to Drosophila, wasps, and honey bees. In Drosophila, regulatory elements in the female-specific exon are bound by Tra/Tra2 proteins and thus signal to the splicing machinery that this particular exon should be included in the final mature mRNA (Hedley and Maniatis 1991; Heinrichs et al. 1998). Short repeats “(U/G)GAAGAU(U/A)” were also found in the female-specific exons of Nasonia vitripennis and honey bee dsx transcripts suggesting that they could act as recognition sites for factors that lead to the activation of female-specific splicing (Bertossa et al. 2009). We also found highly conserved repeats in the female-specific exon of dsx in F. varia (three repeats), M. quadrifasciata (two repeats) and S. postica (three repeats) that share the same core sequence “GAAGAU” with the four repeats found in A. mellifera. These sites may act as regulatory elements that are recognized by fem proteins and lead to the inclusion of the female-specific exon into the mature mRNA of dsx found in females of these species. The number of repeats varies slightly between bee species, possibly indicating differences in the number of proteins that can bind to the female-specific exons of dsx (Figure 4). Additionally, other mechanisms may be involved in the RNA splicing variants of genes of sex determination pathway. For example, in Drosophila, the RNA modification N6-methyladenosine facilitates the female-specific splicing of the primary signal of the sex determination pathway, the gene Sex-lethal (Kan et al. 2017).
Short repeats found in honey bee and stingless bees dsx female-specific exons. a Comparison of short repeats between F. varia, M. quadrifasciata, S. postica, and A. mellifera. Species, architecture of female-specific exons, and repeats at corresponding exons are indicated. Positions of repeats in female exons (gray) are indicated by vertical black stripes. The gray box in the repeat column indicates the core sequence, conserved in all repeats and species analyzed. Information of A. mellifera repeats were obtained from Bertossa et al. (2009)). b Sequence motif identified in dsx female-specific exon of F. varia, M. quadrifasciata, S. postica, and A. mellifera by WebLogo 3.
The sex determination cascade may also play a role in caste differentiation. Levels of female-specific fem transcripts seem to be increased by juvenile hormone in M. interrupta suggesting that fem acts as interaction component between sex and caste determination pathways (Brito et al. 2015). Recent studies pointed to the co-option of the sex determination genes to the nutrition-driven developmental program that leads to dimorphic plasticity in beetles, ants, and honey bees. In honey bees, feminizer is required for the small size polyphenism observed in workers (Roth et al. 2019). In the beetles, it was shown that dsx plays a role in the switch that triggers horn development in males (Zinna et al. 2018). Klein et al. (2016) hypothesized that genes that compose sex determination cascade are able to regulate caste differentiation pathways because these genes evolved to respond to environmental clues (i.e., quantity of food) in addition to the genetic signals (i.e., heterodimers or homo-/hemidimers of Csd protein) that direct sex differentiation. Thus, the investigation of sex determination cascade may shed light on the mechanisms behind the caste differentiation across the diverse group that is the stingless bees.
Our work contributes with the description of sex-specific variants of fem and dsx genes in three stingless bee species. The highly conserved architecture of fem and dsx transcripts and the presence of short repeats in the female-specific exon of dsx suggest conservation of the regulatory mechanisms involved in the sex-specific splicing of these genes.
References
Adams, J., E. D. Rothman, W. E. Kerr and Z. L. Paulino, 1977 Estimation of the number of sex alleles and queen matings from diploid male frequencies in a population of Apis mellifera. Genetics 86: 583–596.
Bertossa, R. C., L. van de Zande and L. W. Beukeboom, 2009 The Fruitless gene in Nasonia displays complex sex-specific splicing and contains new zinc finger domains. Mol. Biol. Evol. 26: 1557–1569.
Beye, M., 2004 The dice of fate: the csd gene and how its allelic composition regulates sexual development in the honey bee, Apis mellifera. Bioessays 26: 1131–1139.
Beye, M., M. Hasselmann, M. K. Fondrk, R. E. Page and S. W. Omholt, 2003 The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114: 419–429.
Boleli, I. C., Z. L. Paulino-Simoes and M. M. Gentile Bitondi, 1999 Cell death in ovarioles causes permanent sterility in Frieseomelitta varia worker bees. J. Morphol. 242: 271–282.
Brito, D. V., C. G. Silva, M. Hasselmann, L. S. Viana, S. Astolfi-Filho et al., 2015 Molecular characterization of the gene feminizer in the stingless bee Melipona interrupta (Hymenoptera: Apidae) reveals association to sex and caste development. Insect Biochem. Mol. Biol. 66: 24–30.
Britten, R. J., and E. H. Davidson, 1969 Gene regulation for higher cells: a theory. Science 165: 349–357.
Burtis, K. C., 2002 Development. Doublesex in the middle. Science 297: 1135–1136.
Cho, S., and P. C Wensink. 1998 Linkage between oligomerization and DNA binding in Drosophila doublesex proteins. Biochemistry 37(32): 11301–11308.
Cho, S., Z. Y. Huang and J. Zhang, 2007 Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177: 1733–1741.
Clark, K., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell and E. W. Sayers, 2016 GenBank. Nucleic Acids Res. 44: D67–72.
Clough, E., E. Jimenez,Y. A. Kim, C. Whitworth, M. C. Neville, et al., 2014 Sex-and tissue-specific functions of Drosophila doublesex transcription factor target genes. Dev. Cell 31(6): 761–773.
Cristino, A. S., A. M. Nascimento, F. Costa Lda and Z. L. Simoes, 2006 A comparative analysis of highly conserved sex-determining genes between Apis mellifera and Drosophila melanogaster. Genet. Mol. Res. 5: 154–168.
de Camargo, C. A., 1979 Sex determination in beesXI Production of diploid males and sex determination in Melipona quadrifasciata. J. Apic. Res. 18: 77–84.
Dzierzon, J., 1845 Gutachten über die von Herrn Direktor Stöhr im ersten und zweiten Kapitel des General-Gutachtens aufgestellten Fragen. Eichstädter Bienenzeitung 1: 119–121.
Erdman, S. E., H. J. Chen, and K. C. Burtis. 1996 Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics 144(4): 1639–1652.
Estoup, A., M. Solignac and J.-m. Cornuet, 1994 Precise assessment of the number of patrilines and of genetic relatedness in honeybee colonies. Proceedings of the Royal Society of LondonSeries B: Biological Sciences 258: 1–7.
Garrett-Engele, C. M., M. L. Siegal, D. S. Manoli, B. C. Williams, H. Li, and B. S. Baker. 2002 intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development, 129(20): 4661–4675.
Gempe, T., M. Hasselmann, M. Schiott, G. Hause, M. Otte et al., 2009 Sex determination in honeybees: two separate mechanisms induce and maintain the female pathway. PLoS Biol. 7: e1000222.
Hartfelder, K., G. R. Makert, C. C. Judice, G. A. Pereira, W. C. Santana et al., 2006 Physiological and genetic mechanisms underlying caste development, reproduction and division of labor in stingless bees. Apidologie 37: 144–163.
Hasselmann, M., T. Gempe, M. Schiott, C. G. Nunes-Silva, M. Otte et al., 2008 Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454: 519–522.
Hedley, M. L., and T. Maniatis, 1991 Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell 65: 579–586.
Heinrichs, V., L. C. Ryner and B. S. Baker, 1998 Regulation of Sex-Specific Selection of fruitless 5′ Splice Sites by transformer and transformer-2. Mol. Cell. Biol. 18: 450–458.
Kan, L., A. V. Grozhik, J. Vedanayagam, D. P. Patil, N. Pang et al., 2017 The m(6) A pathway facilitates sex determination in Drosophila. Nat. Commun. 8: 15737.
Kapheim, K. M., H. Pan, C. Li, S. L. Salzberg, D. Puiu et al., 2015 Genomic signatures of evolutionary transitions from solitary to group living. Science 348: 1139–1143.
Kato, Y., K. Kobayashi, H. Watanabe and T. Iguchi, 2011 Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet. 7: e1001345.
Kearse, M., R. Moir, A. Wilson, S. Stones-Havas, M. Cheung et al., 2012 Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649.
Kerr, W. E., 1974 Sex determination in bees. III. Caste determination and genetic control in Melipona. Insect. Soc. 21: 357–367.
Kerr, W., 1987 Sex determination in bees XXI. Number of xo-heteroalleles in a natural population of Melipona compressipes fasciculata (Apidae). Insect. Soc. 34: 274–279.
Klein, A., Schultner, E., Lowak, H., Schrader, L., Heinze, J., Holman, L., & Oettler, J. 2016. Evolution of social insect polyphenism facilitated by the sex differentiation cascade. PLoS Genet., 12(3), e1005952.
Kumar, S., G. Stecher and K. Tamura, 2016 MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874.
Li, W., A. Cowley, M. Uludag, T. Gur, H. McWilliam et al., 2015 The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43: W580–584.
Marchler-Bauer, A., M. K. Derbyshire, N. R. Gonzales, S. Lu, F. Chitsaz, et al., 2014 CDD: NCBI's conserved domain database. Nucleic Acids Res. 43(D1): D222-D226.
Okonechnikov, K., O. Golosova, M. Fursov and U. team, 2012 Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28: 1166–1167.
Oldroyd, B. P., M. J. Clifton, S. Wongsiri, T. E. Rinderer, H. A. Sylvester et al., 1997 Polyandry in the genus Apis, particularly Apis andreniformis. Behav. Ecol. Sociobiol. 40: 17–26.
Paxton, R. J., L. R. Bego, M. M. Shah and S. Mateus, 2003 Low mating frequency of queens in the stingless bee Scaptotrigona postica and worker maternity of males. Behav. Ecol. Sociobiol. 53: 174–181.
Peters, J. M., D. C. Queller, V. L. Imperatriz–Fonseca, D. W. Roubik and J. E. Strassmann, 1999 Mate number, kin selection and social conflicts in stingless bees and honeybees. Proc. Biol. Sci. 266: 379–384.
Raymond, C., C. Shamu, M. Shen, M. K. J. Seifert, B. Hirsch, et al., 1998 Evidence for evolutionary conservation of sex-determining genes. Nature 391: 691–695. DOI: https://doi.org/10.1038/35618
Roth, A., C. Vleurinck, O. Netschitailo, V. Bauer, M. Otte et al., 2019 A genetic switch for worker nutrition-mediated traits in honeybees. PLoS Biol. 17: e3000171.
Rozen, S., and H. Skaletsky, 2000 Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386.
Shepard, P. J., and K. J. Hertel, 2009 The SR protein family. Genome Biol. 10: 242.
Tóth, E., J. E. Strassmann, P. Nogueira-Neto, V. L. Imperatriz-Fonseca and D. C. Queller, 2002 Male production in stingless bees: variable outcomes of queen–worker conflict. Mol. Ecol. 11: 2661–2667.
Viana, B. F., J. G. da Encarnação Coutinho, L. A. Garibaldi, B. Gastagnino, G. Laercio et al., 2014 Stingless bees further improve apple pollination and production. Journal of Pollination Ecology 14.
Vollet-Neto, A., S. Koffler, C. dos Santos, C. Menezes, F. Nunes et al., 2018 Recent advances in reproductive biology of stingless bees. Insect. Soc. 65: 201–212.
Wilkins, A. S., 1995 Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17: 71–77.
Woyke, J., 1965 Genetic proof of the origin of drones from fertilized eggs of the honeybee. J. Apic. Res. 4: 7–11.
Zinna, R. A., H. Gotoh, T. Kojima and T. Niimi, 2018 Recent advances in understanding the mechanisms of sexually dimorphic plasticity: insights from beetle weapons and future directions. Curr. Opin. Insect Sci. 25: 35–41.
Acknowledgments
The authors thank Prof. Dr. Isabel dos Santos and Dr. Sheina Koffler (University of São Paulo, São Paulo, Brazil) for providing the male samples of M. quadrifasciata, Ivan de Castro and Jairo de Souza (University of São Paulo, Ribeirão Preto, Brazil) for giving support to collect samples of F. varia and M. quadrifasciata.
Funding
This research was financially supported by FAPESP (Project number 2016/06657-0), MCTIC/CNPq/Universal 14/2014 (Process number 454103/2014-0), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES) (Finance Code 001).
Author information
Authors and Affiliations
Contributions
FF, JB performed the experiments and data analysis. ZS acquired funding. FF, AC, FN, ZS wrote the manuscript. ZS, AC conceived and supervised the research. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Manuscript editor: Klaus Hartfelder
Caractérisation des variantes sexospécifiques des gènes doublesex et féminisant chez les espèces d'abeilles sans dard.
Abeilles sans dard / détermination du sexe / Melipona quadrifasciata / Scaptotrigona postica / Frieseomelitta varia .
Charakterisierung von geschlechtsspezifischen Varianten der doublesex- und feminizer-Gene bei Stachellosen Bienen.
Stachellose Bienen/ Geschlechtsbestimmung/ Melipona quadrifasciata / Scaptotrigona postica / Frieseomelitta varia .
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 572 kb)
Rights and permissions
About this article
Cite this article
de Paula Freitas, F.C., Buchholz, J., Nunes, F.M.F. et al. Characterization of sex-specific variants of doublesex and feminizer genes in stingless bee species. Apidologie 51, 469–480 (2020). https://doi.org/10.1007/s13592-020-00735-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-020-00735-8