Abstract
Undifferentiated pleomorphic sarcoma (UPS), previously termed malignant fibrous histiocytoma, is one of the most aggressive sarcomas with no identifiable line of differentiation. Although the molecular mechanism of oncogenesis in UPS has not been clarified, radiation exposure is considered to be a risk factor in the development of UPS. In the treatment of UPS, surgical treatment remains the most important modality. While chemotherapy is considered in unresectable or metastatic cases, UPS is known to be refractory to conventional chemotherapy, leading to an unfavorable prognosis. To improve the clinical outcome of this condition, novel treatment methods are urgently needed. Patient-derived cell lines are essential tools in preclinical studies. However, owing to the rarity of UPS, only four UPS cell lines are publicly available. Thus, we established a novel UPS cell line, NCC-UPS3-C1, using a surgically resected tumor from a patient with radiation-associated UPS. NCC-UPS3-C1 cells had multiple genomic deletions including the tumor suppressor genes CDKN2A and CDKN2B. NCC-UPS3-C1 cells demonstrated constant growth, spheroid formation, and aggressive invasion ability. We also conducted a screening test using 214 drugs and identified that the histone deacetylase inhibitor, romidepsin, is highly effective on NCC-UPS3-C1 cells. Thus, we concluded that the NCC-UPS3-C1 cell line is a useful tool in preclinical studies for UPS.




Similar content being viewed by others
References
WHO classification of tumours of soft tissue and bone. 5th edn. Lyon: IARC; 2020.
Widemann BC, Italiano A. Biology and management of undifferentiated pleomorphic sarcoma, myxofibrosarcoma, and malignant peripheral nerve sheath tumors: state of the art and perspectives. J Clin Oncol. 2018;36:160–7.
Chen S, Huang W, Luo P, et al. Undifferentiated pleomorphic sarcoma: long-term follow-up from a large institution. Cancer Manag Res. 2019;11:10001–9.
Gladdy RA, Qin LX, Moraco N, et al. Do radiation-associated soft tissue sarcomas have the same prognosis as sporadic soft tissue sarcomas? J Clin Oncol. 2010;28:2064–9.
Dineen SP, Roland CL, Feig R, et al. Radiation-associated undifferentiated pleomorphic sarcoma is associated with worse clinical outcomes than sporadic lesions. Ann Surg Oncol. 2015;22:3913–20.
Joo MW, Kang YK, Ogura K, et al. Post-radiation sarcoma: a study by the Eastern Asian Musculoskeletal Oncology Group. PLoS ONE. 2018;13:e0204927.
Callesen LB, Safwat A, Rose HK, Sorensen FB, Baad-Hansen T, Aggerholm-Pedersen N. Radiation-induced sarcoma: a retrospective population-based study over 34 years in a single institution. Clin Oncol (R Coll Radiol). 2021;33:e232–8.
Vodanovich DA, Spelman T, May D, Slavin J, Choong PFM. Predicting the prognosis of undifferentiated pleomorphic soft tissue sarcoma: a 20-year experience of 266 cases. ANZ J Surg. 2019;89:1045–50.
Kresse SH, Ohnstad HO, Bjerkehagen B, Myklebost O, Meza-Zepeda LA. DNA copy number changes in human malignant fibrous histiocytomas by array comparative genomic hybridisation. PLoS ONE. 2010;5:e15378.
Carneiro A, Francis P, Bendahl PO, et al. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: different sides of a single coin? Lab Invest. 2009;89:668–75.
Simons A, Schepens M, Jeuken J, et al. Frequent loss of 9p21 (p16(INK4A)) and other genomic imbalances in human malignant fibrous histiocytoma. Cancer Genet Cytogenet. 2000;118:89–98.
Perot G, Chibon F, Montero A, et al. Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics. Am J Pathol. 2010;177:2080–90.
Gibault L, Perot G, Chibon F, et al. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol. 2011;223:64–71.
Kamat NV, Million L, Yao DH, et al. The outcome of patients with localized undifferentiated pleomorphic sarcoma of the lower extremity treated at Stanford University. Am J Clin Oncol. 2019;42:166–71.
Goldblum JR. An approach to pleomorphic sarcomas: can we subclassify, and does it matter? Mod Pathol. 2014;27(Suppl 1):S39-46.
Grimer R, Judson I, Peake D, Seddon B. Guidelines for the management of soft tissue sarcomas. Sarcoma. 2010;2010:506182.
Reichardt P. Soft tissue sarcomas, a look into the future: different treatments for different subtypes. Future Oncol. 2014;10:s19-27.
Saito S, Morita K, Kohara A, et al. Use of BAC array CGH for evaluation of chromosomal stability of clinically used human mesenchymal stem cells and of cancer cell lines. Hum Cell. 2011;24:2–8.
Ben-David U, Beroukhim R, Golub TR. Genomic evolution of cancer models: perils and opportunities. Nat Rev Cancer. 2019;19:97–109.
Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61.
Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23.
Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61.
Goodspeed A, Heiser LM, Gray JW, Costello JC. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res. 2016;14:3–13.
Hattori E, Oyama R, Kondo T. Systematic review of the current status of human sarcoma cell lines. Cells. 2019;8:157.
Bairoch A. The cellosaurus, a cell-line knowledge resource. J Biomol Tech. 2018;29:25–38.
Amm HM, DeVilliers P, Srivastava AR, Diniz MG, Siegal GP, MacDougall M. Mandibular undifferentiated pleomorphic sarcoma: molecular analysis of a primary cell population. Clin Exp Dent Res. 2020;6:495–505.
De Vita A, Recine F, Mercatali L, et al. Primary culture of undifferentiated pleomorphic sarcoma: molecular characterization and response to anticancer agents. Int J Mol Sci. 2017;18:2662.
Tsuchiya R, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of NCC-SS4-C1: a novel patient-derived cell line of synovial sarcoma. Hum Cell. 2021;34:998–1007.
Sin Y, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of a novel alveolar rhabdomyosarcoma cell line, NCC-aRMS1-C1. Hum Cell. 2020;33:1311–20.
Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–5.
Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Mizusawa H. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tiss Cult Res Commun. 1999;18:329–38.
Capes-Davis A, Reid YA, Kline MC, et al. Match criteria for human cell line authentication: where do we draw the line? Int J Cancer. 2013;132:2510–9.
Bui NQ, Przybyl J, Trabucco SE, et al. A clinico-genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res. 2019;9:12.
Orth MF, Gerke JS, Knosel T, et al. Functional genomics identifies AMPD2 as a new prognostic marker for undifferentiated pleomorphic sarcoma. Int J Cancer. 2019;144:859–67.
De Vita A, Recine F, Miserocchi G, et al. The potential role of the extracellular matrix in the activity of trabectedin in UPS and L-sarcoma: evidences from a patient-derived primary culture case series in tridimensional and zebrafish models. J Exp Clin Cancer Res. 2021;40:165.
Takai Y, Oyama R, Kito F, et al. Establishment and characterizationof cell line of undifferentiated pleomorphic sarcoma. Tiss Cult Res Commun. 2017;36:41–8.
Iyer SP, Foss FF. Romidepsin for the treatment of peripheral T-cell lymphoma. Oncologist. 2015;20:1084–91.
Becker M, Graf C, Tonak M, et al. Xenograft models for undifferentiated pleomorphic sarcoma not otherwise specified are essential for preclinical testing of therapeutic agents. Oncol Lett. 2016;12:1257–64.
Saitoh Y, Bureta C, Sasaki H, et al. The histone deacetylase inhibitor LBH589 inhibits undifferentiated pleomorphic sarcoma growth via downregulation of FOS-like antigen 1. Mol Carcinog. 2019;58:234–46.
Rivera-Reyes A, Ye S, Gloria EM, et al. YAP1 enhances NF-kappaB-dependent and independent effects on clock-mediated unfolded protein responses and autophagy in sarcoma. Cell Death Dis. 2018;9:1108.
Acknowledgements
We thank Drs. E. Kobayashi, S. Iwata, S. Fukushima, M. Nakagawa, S. Ozaki, C. Sato (Department of Musculoskeletal Oncology), Dr. T. Ushikusa (Department of Diagnostic Pathology), and the National Cancer Center Hospital, for sampling tumor tissue specimens from surgically resected materials. We also appreciate the technical assistance provided by Ms. Y. Kuwata (Division of Rare Cancer Research). We appreciate the technical support provided by Ms. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would like to thank Editage (www.editage.jp) for their help with English language editing and constructive comments on the manuscript. This research was received technical assistance from the Fundamental Innovative Oncology Core at the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (Grant Number 20ck0106537h0001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The ethical committee of the National Cancer Center approved the use of clinical materials for this study (approval number 2004-050).
Informed consent
Written informed consent for publication was provided by the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13577_2021_633_MOESM8_ESM.tif
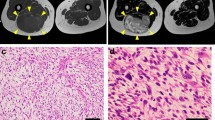
Supplementary file8 Supplementary Fig. 3 Tumorigenesis in nude mice. (A) NCC-UPS3-C1 cells transplanted into BALB/c nude mice formed a small tumor mass under the described condition. The yellow circles indicate the tumor. (B) The H&E-stained tumor represents the dense proliferation of pleomorphic cells. (C) The graph showing estimated tumor volume. Bars represent the mean standard error (TIF 1337 KB)
13577_2021_633_MOESM9_ESM.tif
Supplementary file9 Supplementary Fig. 4 Growth curves for the IC50 value calculations of 24 anticancer agents (TIF 618 KB)
Rights and permissions
About this article
Cite this article
Tsuchiya, R., Yoshimatsu, Y., Noguchi, R. et al. Establishment and characterization of NCC-UPS3-C1: a novel patient-derived cell line of undifferentiated pleomorphic sarcoma. Human Cell 35, 384–391 (2022). https://doi.org/10.1007/s13577-021-00633-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00633-w