Abstract
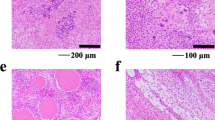
Pleomorphic sarcoma (PS) is a heterogeneous group of malignant mesenchymal tumors without a specific histological lineage of differentiation. PS is genetically characterized by genetic instability and diversity and histologically characterized by morphological pleomorphism. PS is one of the most common soft tissue sarcomas. The only curative treatment for PS is complete surgical resection, in which neoadjuvant radiotherapy is frequently combined. PS demonstrates both local recurrence and metastasis after surgical treatment, and effective systemic chemotherapy has not yet been established. Patient-derived cancer cell lines are critical tools for basic and preclinical studies in the development of chemotherapy. However, only six PS cell lines are available from the public cell bank, and none of them are derived from PS after neoadjuvant radiotherapy, despite the fact that radiotherapy causes changes in the posttreatment cancer genome. Here, we reported a novel cell line of PS from a primary tumor specimen resected after neoadjuvant radiotherapy and named it NCC-PS1-C1. NCC-PS1-C1 cells showed a variety of copy number alterations and pathological mutations in TP53. NCC-PS1-C1 cells demonstrated constant proliferation, spheroid formation, and invasion capability in vitro. The screening of antitumor agents in NCC-PS1-C1 cells showed that bortezomib and romidepsin were effective against PS. In conclusion, we report a novel PS cell line from a primary tumor resected after neoadjuvant radiotherapy. We believe that NCC-PS1-C1 will be a useful tool for the development of novel chemotherapies for PS, especially for recurrent cases after neoadjuvant radiotherapy.




Similar content being viewed by others
References
Carvalho SD, Pissaloux D, Crombé A, Coindre JM, Le Loarer F. Pleomorphic sarcomas: the state of the art. Surg Pathol Clin. 2019;12:63–105.
Hornick JL. Subclassification of pleomorphic sarcomas: How and why should we care? Ann Diagn Pathol. 2018;37:118–24.
Toulmonde M, Lucchesi C, Verbeke S, et al. High throughput profiling of undifferentiated pleomorphic sarcomas identifies two main subgroups with distinct immune profile, clinical outcome and sensitivity to targeted therapies. EBioMedicine. 2020;62: 103131.
Chen S, Huang W, Luo P, et al. Undifferentiated pleomorphic sarcoma: long-term follow-up from a large institution. Cancer Manag Res. 2019;11:10001–9.
Weiss SW, Enzinger FM. Malignant fibrous histiocytoma: an analysis of 200 cases. Cancer. 1978;41:2250–66.
Dei Tos AP. Classification of pleomorphic sarcomas: where are we now? Histopathology. 2006;48:51–62.
Wakely PE Jr. Cytopathology of myxofibrosarcoma: a study of 66 cases and literature review. J Am Soc Cytopathol. 2021;10:300–9.
Gervais MK, Callegaro D, Gronchi A. The evolution of adjuvant/neoadjuvant trials for resectable localized sarcoma. J Surg Oncol. 2022;125:17–27.
Keung EZ, Tsai JW, Ali AM, et al. Analysis of the immune infiltrate in undifferentiated pleomorphic sarcoma of the extremity and trunk in response to radiotherapy: rationale for combination neoadjuvant immune checkpoint inhibition and radiotherapy. Oncoimmunology. 2018;7: e1385689.
Pisters PW, Harrison LB, Leung DH, Woodruff JM, Casper ES, Brennan MF. Long-term results of a prospective randomized trial of adjuvant brachytherapy in soft tissue sarcoma. J Clin Oncol. 1996;14:859–68.
Weitz J, Antonescu CR, Brennan MF. Localized extremity soft tissue sarcoma: improved knowledge with unchanged survival over time. J Clin Oncol. 2003;21:2719–25.
Vodanovich DA, Spelman T, May D, Slavin J, Choong PFM. Predicting the prognosis of undifferentiated pleomorphic soft tissue sarcoma: a 20-year experience of 266 cases. ANZ J Surg. 2019;89:1045–50.
Huang RX, Zhou PK. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct Target Ther. 2020;5:60.
Tentler JJ, Tan AC, Weekes CD, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–50.
Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–6.
Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–84.
Goodspeed A, Heiser LM, Gray JW, Costello JC. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res. 2016;14:3–13.
Hideyuki T, Yoko T, Daisuke M, Miharu K, Tohru M, Hiroshi M. Cell line individualization by str multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tissue Cult Res Commun. 1999;18:329–38.
Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci USA. 2001;98:8012–7.
Drexler HG, Dirks WG, MacLeod RA, Uphoff CC. False and mycoplasma-contaminated leukemia-lymphoma cell lines: time for a reappraisal. Int J Cancer. 2017;140:1209–14.
Yoshimatsu Y, Noguchi R, Tsuchiya R, et al. Establishment and characterization of NCC-CDS2-C1: a novel patient-derived cell line of CIC-DUX4 sarcoma. Hum Cell. 2020;33:427–36.
Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–7.
Sunami K, Ichikawa H, Kubo T, et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: a hospital-based study. Cancer Sci. 2019;110:1480–90.
Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–5.
Mohseny AB, Machado I, Cai Y, et al. Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Lab Invest. 2011;91:1195–205.
Klijn C, Durinck S, Stawiski EW, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–12.
Dutil J, Chen Z, Monteiro AN, Teer JK, Eschrich SA. An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines. Cancer Res. 2019;79:1263–73.
Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503–8.
Cox C, Teknos TN, Barrios M, Brewer GJ, Dick RD, Merajver SD. The role of copper suppression as an antiangiogenic strategy in head and neck squamous cell carcinoma. Laryngoscope. 2001;111:696–701.
Li GZ, Okada T, Kim YM, et al. Rb and p53-deficient myxofibrosarcoma and undifferentiated pleomorphic sarcoma require Skp2 for survival. Cancer Res. 2020;80:2461–71.
Kocakavuk E, Anderson KJ, Varn FS, et al. Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer. Nat Genet. 2021;53:1088–96.
Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–6.
Carneiro A, Francis P, Bendahl PO, et al. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: different sides of a single coin? Lab Invest. 2009;89:668–75.
Oyama R, Kito F, Sakumoto M, et al. Establishment and proteomic characterization of a novel cell line, NCC-UPS2-C1, derived from a patient with undifferentiated pleomorphic sarcoma. In Vitro Cell Dev Biol Anim. 2018;54:257–63.
Tsuchiya R, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of NCC-UPS3-C1: a novel patient-derived cell line of undifferentiated pleomorphic sarcoma. Hum Cell. 2022;35:384–91.
Ono T, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of NCC-UPS4-C1: a novel cell line of undifferentiated pleomorphic sarcoma from a patient with Li-Fraumeni syndrome. Hum Cell. 2022;35:756–66.
Kito F, Oyama R, Sakumoto M, et al. Establishment and characterization of a novel cell line, NCC-MFS1-C1, derived from a patient with myxofibrosarcoma. Hum Cell. 2019;32:214–22.
Noguchi R, Yoshimatsu Y, Ono T, et al. Establishment and characterization of NCC-MFS2-C1: a novel patient-derived cancer cell line of myxofibrosarcoma. Hum Cell. 2021;34:246–53.
Tsuchiya R, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of NCC-MFS3-C1: a novel patient-derived cell line of myxofibrosarcoma. Hum Cell. 2021;34:1266–73.
Yoshimatsu Y, Noguchi R, Tsuchiya R, et al. Establishment and characterization of NCC-MFS4-C1: a novel patient-derived cell line of myxofibrosarcoma. Hum Cell. 2021;34:1911–8.
Tsuchiya R, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of NCC-MFS5-C1: a novel patient-derived cell line of myxofibrosarcoma. Cells. 2022;11:207.
Liu J, Zhao R, Jiang X, Li Z, Zhang B. Progress on the application of bortezomib and bortezomib-based nanoformulations. Biomolecules. 2021;12:51.
Li CF, Wang JM, Kang HY, et al. Characterization of gene amplification-driven SKP2 overexpression in myxofibrosarcoma: potential implications in tumor progression and therapeutics. Clin Cancer Res. 2012;18:1598–610.
Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. 2017;18:1414.
Saitoh Y, Bureta C, Sasaki H, et al. The histone deacetylase inhibitor LBH589 inhibits undifferentiated pleomorphic sarcoma growth via downregulation of FOS-like antigen 1. Mol Carcinog. 2019;58:234–46.
Acknowledgements
We thank Drs. F. Nakatani, S. Iwata, S. Fukushima, T. Komatsubara (Department of Musculoskeletal Oncology), Drs. C. Sato, H. Tanaka, T. Shibayama (Department of Diagnostic Pathology) and the National Cancer Center Hospital for sampling tumor tissue specimens from surgically resected materials. We also appreciate the technical assistance provided by Mrs. Y. Kuwata (Division of Rare Cancer Research), and the technical support provided by Mrs. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would like to thank Editage (www.editage.jp) for providing English language editing services and for their constructive comments on the manuscript. This research was technically assisted by the Fundamental Innovative Oncology Core in the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (grant number: 20ck0106537h0002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
The Ethical Committee of the National Cancer Center approved the use of clinical materials for this study (approval number 2004–050). Animal experiments were conducted in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute.
Informed consent
Written informed consent was provided by the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13577_2022_787_MOESM1_ESM.tiff
Supplementary file1 Supplementary Fig. 1 Short tandem repeat patterns of NCC-PS1-C1 (passage 21) and original tumor tissue. (a) Short tandem repeat patterns of NCC-PS1-C1 and (b) short tandem repeat patterns of original tumor tissue of NCC-PS1-C1 (TIFF 3923 KB)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Akiyama, T., Yoshimatsu, Y., Noguchi, R. et al. Establishment and characterization of NCC-PS1-C1: a novel cell line of pleomorphic sarcoma from a patient after neoadjuvant radiotherapy. Human Cell 35, 2011–2019 (2022). https://doi.org/10.1007/s13577-022-00787-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00787-1