Abstract
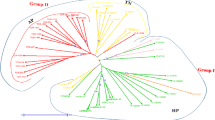
Sugarcane is a crop of economic importance providing sugar and bioenergy resource which has a substantial contribution to the national GDP of India as well as other countries. Genomes of the several grasses have co-evolved which share large-scale synteny and genome collinearity; hence it is possible to employ EST-SSR markers of one grass species for genetic study of other related species. A set of sixty potentially informative EST-derived microsatellite (SSR) markers were employed to examine their cross-taxa transferability across the limit of taxonomic boundaries of different grass genomes. The high (96.6%) rate of SSRs cross-transferability observed in genus Saccharum followed by Erianthus (93.3%), Narenga (88.3%), Sclerostachya (86.3%), and Miscanthus (83.3%). Relatively lower rates of cross amplification were observed in other cereal crops including Sorghum bicolor, Pennisetum glaucum, Zea mays, Triticum aestivum, Oryza sativa, and Avena sativa. Cross-transferable EST-SSR markers were utilized to evaluate phylogenetic relationships among genus Saccharum, closely related genera of Saccharum complex, and other allied cereal grasses. The polymorphic information content values ranged from 0.35 to 0.95, with an average of 0.71. Such a high rates of cross-taxa transferability of the EST-SSRs make them a potent tool for genetic studies and comparative genomics across wide evolutionary distances within and across genera along with technologically simplicity and cost effectiveness. EST-SSRs will facilitate to devise potent tools for genetic improvement through advance molecular breeding of such crops for high grain productivity and better quality. They will especially be useful for the orphan grass species, which contribute a major part of the global food security.




Similar content being viewed by others
Abbreviations
- EST-SSRs:
-
Expressed sequence tags derived simple sequence repeats
- FAOSTAT:
-
Food and Agriculture Organization of the United Nations-Statistics Division
- 5′ UTR:
-
Five prime untranslated regions
- 3′ UTR:
-
Three prime untranslated regions
- hnRNA:
-
Heterogeneous nuclear RNA
- MISA:
-
MIcroSAtellite Identification Tool
References
Aggarwal RK, Prasad SH, Varshney RK, Prasanna R, Bhat KV, Singh L (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet 114:359–372
Aitken KS, Jackson PA, McIntyre CL (2005) A combination of AFLP and SSR markers provides extensive map coverage and identification of homo(eo)logous linkage groups in a sugarcane cultivar. Theor Appl Genet 110:789–801
Bennetzen JL, Freeling M (1993) Grasses as a single genetic system: genome composition, co-llinearity and compatibility. Trends Genet 9:259–261
Besse P, McIntyre CL, Berding N (1997) Characterisation of Erianthus sect. Ripidium and Saccharum germplasm (Andropogoneae-Saccharinae) using RFLP markers. Euphytica 93:283–292
Brandes EW (1958) Origin, classification and characteristics. In: Artschwager E, Brandes EW (eds) Sugarcane (Saccharum officinarum L.), vol 122. US department of agriculture handbook. U.S. Government Print Office, Washington, D.C, pp 1–35
Burnquist WL, Sorrells ME, Tanksley SD (1995) Characterization of genetic variability in Saccharum germplasm by means of Restriction Fragment Length Polymorphism (RFLP) analysis. Proc Int Soc Sugarcane Technol 21:355–365
Castillo A, Budak H, Varshney RK, Dorado G, Graner A, Hernandez P (2008) Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis in Hordeum chilense. BMC Plant Biol 8:97. https://doi.org/10.1186/1471-2229-8-97
Chagné D, Chaumeil P, Ramboer A, Collada C, Guevara A, Cervera MT, Vendramin GG, Garcia V, Frigerio JM, Echt C, Richardson T (2004) Cross-species transferability and mapping of genomic and cDNA SSRs in pines. Theor Appl Genet 109:1204–1214
Chandra A, Jain R, Soloman S, Srivastava S, Roy AK (2013) Exploiting EST databases for the development and characterisation of 3425 gene-tagged CISP markers in biofuel crop sugarcane and their transferability in cereals and orphan tropical grasses. BMC Res Notes 6:47
Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor Appl Genet 100:713–722
Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to Erianthus and sorghum. Plant Sci 160:1115–1123
Daniels J, Smith P, Panton N, Williams CA (1975) The origin of the genus Saccharum. Sugarcane Breed. Newsl 36:24–39
Dayanandan S, Bawa KS, Kesseli R (1997) Conservation of microsatellites among tropical trees (Leguminosae). Am J Bot 84:1658–1663
D’Hont A, Glaszman JC (2001) Sugarcane genome analysis with molecular markers, a first decade of research. Proc Int Soc Sugarcane Technol 24:556–559
Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A, Arús P, Laigret F (2002) Development of microsatellite markers in peach (Prunus persica L. Batsch) and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L). Theor Appl Genet 105:127–138
Disasa T, Feyissa T, Sertse D (2016) Transferability of sorghum microsatellite markers to bamboo and detection of polymorphic markers. Open Biotech J 10:223–233
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dufour P, Grivet L, D’Hont A, Deu M, Trouche G, Glaszmann JC, Hamon P (1996) Comparative genetic mapping between duplicated segments on maize chromosomes 3 and 8 and homoeologous regions in sorghum and sugarcane. Theor Appl Genet 92:1024–1030
FAOSTAT (2014) Food and Agricultural Organisation of the United Nations. http://faostat.fao.org/site/291/default.aspx. Accessed 30 Apr 2018
Feltus FA, Singh HP, Lohithaswa HC, Schulze SR, Silva TD, Paterson AH (2006) A comparative genomics strategy for targeted discovery of single-nucleotide polymorphisms and conserved-noncoding sequences in orphan crops. Plant Physiol 140:1183–1191
Feuillet C, Keller B (1999) High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA 96:82658270
Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95:1971–1974
Gao LZ, Zhang CH, Jia JZ (2005) Cross-species transferability of rice microsatellites in its wild relatives and the potential for conservation genetic studies. Genet Resour Crop Evol 52:931–940
Gaut BS, Doebley JF (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94:6809–6814
Grivet L, Arruda P (2002) Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr Opin Plant Biol 5:122–127
Hampl V, Pavlicek A, Flegr J (2001) Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with a freeware program FreeTree: application to trichomonad parasites. Int J Syst Evol Microbiol 51:731–735
Hancock JM (1999) Microsatellites and other simple sequences: genomic context and mutational mechanisms. In: Goldstein DB, Schlötterer C (eds) Microsatellites: evolution and applications. Oxford University Press, Oxford
Ilic K, SanMiguel PJ, Bennetzen JL (2003) A complex history of rearrangement in and orthologous region of the maize, sorghum, and rice genomes. Proc Natl Acad Sci USA 100(21):12265–12270
Jarne P, Lagoda PJL (1996) Microsatellites from molecules to population and back. Trends Mol Ecol 11:424–429
Kandel R, Singh HP, Singh BP, Harris-Shultz KR, Anderson WF (2016) Assessment of genetic diversity in Napier grass (Pennisetum purpureum Schum.) using microsatellite, single-nucleotide polymorphism and insertion-deletion markers from pearl millet (Pennisetum glaucum L). Plant Mol Biol Rep 34:265–272
Kantety RV, Rota ML, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kuleung C, Baenziger PS, Dweikat I (2004) Transferability of SSR markers among wheat, rye, and triticale. Theor Appl Genet 108:1147–1150
Kumar R, Appunu C, Anna Durai A, Premachandran MN, Viola VR, Ram B, Mahadevaiah C, Meena MR, Manjunatha T (2015) Genetic confirmation and field performance comparison for yield and quality among advanced generations of Erianthus arundinaceus, E. bengalense and Saccharum spontaneum cyto-nuclear genome introgressed sugarcane intergeneric hybrids. Sugar Tech 17:379–385
Kumar S, Parekh MJ, Patel CB, Zala HN, Sharma R, Kulkarni KS, Fougat RS, Bhatt RK, Sakure AA (2016) Development and validation of EST-derived SSR markers and diversity analysis in cluster bean (Cyamopsis tetragonoloba). J Plant Biochem Biotech 25:263–269
Kurata N, Moore G, Nagamura Y, Foote T, Yano M, Minobe Y, Gale M (1994) Conservation of genome structure between rice and wheat. Biotechnology 12:276–278
Li YC, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007
Li L, Wang J, Guo Y, Jiang F, Xu Y, Wang Y, Pan H, Han G, Li R, Li S (2008) Development of SSR markers from ESTs of gramineous species and their chromosome location on wheat. Prog Nat Sci 18:1485–1490
Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J (2000) An optimized protocol for analysis of EST sequences. Nucleic Acids Res 28:3657–3665
Litt M, Luty JA (1989) A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Human Genet 44:397–401
Lorieux M, Rami JF, Glaszmann JC, Grivet L (2001) A direct comparison between the genetic maps of sorghum and rice. Theor Appl Genet 102:379–386
Milbourne D, Meyer R, Bradshaw JE, Baird E, Bonar N, Povan J, Powell W, Waugh R (1997) Comparison of PCR-based marker system the analysis of genetic relationships in cultivated potato. Mol Breed 3:127–136
Nair NV, Nair S, Sreenivasan TV, Mohan M (1999) Analysis of genetic diversity and phylogeny in Saccharum and related genera using RAPD markers. Genet Resour Crop Evol 46:73–79
Oliveira KM, Pinto LR, Marconi TG, Mollinari M, Ulian EC, Chabregas SM, Falco MC, Burnquist W, Garcia AA, Souza AP (2009) Characterization of new polymorphic functional markers for sugarcane. Genome 52:191–209
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. J Comp Appl Biosc 12:357–358
Pandey G, Misra G, Kumari K, Gupta SA, Parida SK, Chattopadhyay DE, Prasad MA (2013) Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res 20:197–207
Parekh MJ, Kumar S, Zala HN, Fougat RS, Patel CB, Bosamia TC, Kulkarni KS, Parihar A (2016) Development and validation of novel fiber relevant dbEST–SSR markers and their utility in revealing genetic diversity in diploid cotton (Gossypium herbaceum and G. arboreum). Ind Crops Prod 83:620–629
Parida SK, Rajkumar KA, Dalal V, Singh NK, Mohapatra T (2006) Unigene derived microsatellite markers for the cereal genomes. Theor Appl Genet 112:808–817
Parida SK, Kalia SK, Kaul S, Dalal V, Hemaprabha G, Selvi A, Pandit A, Singh A, Gaikwad K, Sharma TR, Srivastava PS (2009) Informative genomic microsatellite markers for efficient genotyping applications in sugarcane. Theor Appl Genet 118:327–338
Parida SK, Pandit A, Gaikwad K, Sharma TR, Srivastava PS, Singh NK, Mohapatra T (2010) Functionally relevant microsatellites in sugarcane unigenes. BMC Plant Biol 10:251
Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidizaion predating divergence of the cereals and its consequences for comparative genomics. Proc Natl Acad Sci 101:9903–9908
Pinto LR, Oliveira KM, Marconi T, Garcia AA, Ulian EC, De Souza AP (2006) Characterization of novel sugarcane expressed sequence tag microsatellites and their comparison with genomic SSRs. Plant Breed 125:378–384
Powell W, Morgante R, Andre C, Hanafey M, Vogel J, Tingey S, Rafalsky A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Ramu P, Kassahun B, Senthilvel S, Kumar AC, Jayashree B, Folkertsma RT, Ananda Reddy L, Kuruvinashetti MS, Haussmann BIG, Hash CT (2009) Exploiting rice-sorghum synteny for targeted development of EST-SSRs to enrich the sorghum genetic linkage map. Theor Appl Genet 119:1193–1204
Saha MC, Mian MAR, Eujayl I (2004) Tall fescue EST-SSR markers with transferability across several grass species. Theor Appl Genet 109:783–791
Selvi A, Nair NV, Balasundaram MT (2003) Evaluation of maize microsatellite markers for genetic diversity analysis and fingerprinting in sugarcane. Genome 46:394–403
Selvi A, Nair NV, Noyer JL, Singh NK, Balasundaram N, Bansal KC, Koundal KR, Mohapatra T (2006) AFLP analysis of the phonetic organization and genetic diversity in the sugarcane complex, Saccharum and Erianthus. Genet Resour Crop Evol 53:831–842
Selvi A, Mukunthan N, Shanthi RM, Govindaraj P, Singaravelu B, Karthik Prabu T (2008) Assessment of genetic relationships and marker identification in sugarcane cultivars with different levels of top borer resistance. Sugar Tech 10:53–59
Sharma RK, Gupta P, Sharma V, Sood A, Mohapatra T, Ahuja PS (2008) Evaluation of rice and sugarcane SSR markers for phylogenetic and genetic diversity analyses in bamboo. Genome 51:91–103
Sim SC, Yu JK, Jo Y, Sorrells ME, Jung G (2009) Transferability of cereal EST-SSR markers to ryegrass. Genome 52:431–437
Singh NK, Raghuvanshi S, Srivastava SK, Gaur A, Pal AK et al (2004) Sequence analysis of the long arm of rice chromosome 11 for rice-wheat synteny. Funct Integr Genomics 4:102–117
Singh RK, Singh RB, Singh SP, Sharma ML (2011) Identification of sugarcane microsatellites associated to sugar content in sugarcane and transferability to other cereal genomes. Euphytica 182:335–354
Singh RK, Singh RB, Singh SP, Sharma ML (2012) Genes tagging and molecular diversity of red rot susceptible/tolerant sugarcane cultivars using c-DNA and unigene derived markers. World J Microbiol Biotech 28:1669–1679
Singh RK, Jena SN, Khan S, Yadav S, Banarjee N, Raghuvanshi S, Bhardwaj V, Dattamajumder SK, Kapur R, Solomon S, Swapna M (2013a) Development, cross-species/genera transferability of novel EST-SSR markers and their utility in revealing population structure and genetic diversity in sugarcane. Gene 524:309–329
Singh RK, Singh RB, Singh SP, Mishra N, Rastogi J, Sharma ML, Kumar A (2013b) Genetic diversity among the Saccharum spontaneum clones and commercial hybrids through SSR markers. Sugar Tech 15:109–115
Singh RB, Srivastva S, Verma AK, Singh B, Singh RK (2014a) Importance and progress of microsatellite markers in Sugarcane (Saccharum spp. hybrids). Ind J Sugar Techno 29:1–12
Singh RB, Srivastava S, Rastogi J, Gupta GN, Tiwari NN, Singh B, Singh RK (2014b) Molecular markers: exploited in crop improvement practices. Res Environ Life Sci 7:223–232
Singh RB, Singh B, Singh RK (2015) Development of microsatellite (SSRs) markers and evaluation of genetic variability within sugarcane commercial varieties (Saccharum spp. hybrids). Int J Adv Res 3:700–708
Singh RB, Singh B, Singh RK (2017) Study of genetic diversity of sugarcane (Saccharum) species and commercial varieties through TRAP molecular markers. Ind J Plant Physiol 22:332–338
Singh RB, Singh B, Singh RK (2018) Evaluation of genetic diversity in Saccharum species clones and commercial varieties employing molecular (SSRs) and physiological markers. Ind J Plant Genet Resour 31:17–26
Singh RB, Singh B, Singh RK (2019) Development of potential dbEST-derived microsatellite markers for genetic evaluation of sugarcane and related cereal grasses. Ind Crops Prod 128:38–47
Sreenivasan TV, Ahloowalia BS, Heinz DJ (1987) Cytogenetics. In: Heinz DJ (ed) Sugarcane improvement through breeding. Elsevier, Amsterdam
Suman A, Kimbeng CA, Edme SJ, Veremis J (2008) Sequence-related amplified polymorphism (SRAP) markers for assessing genetic relationships and diversity in sugarcane germplasm collections. Plant Gene Resour 6:222–231
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Bio Lett 7:537–546
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotech 23:48–55
Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH (1989) Data of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86:6201–6205
Zala HN, Kulkarni KS, Bosamia TC, Shukla YM, Kumar S, Fougat RS, Patel A (2017) Development and validation of EST derived SSR markers with relevance to downy mildew (Sclerospora graminicola Sacc.) resistance in pearl millet [Pennisetum glaucum (L.) R. Br.]. J Plant Biochem Biotech 26:356–365
Zeid M, Yu JK, Goldowitz I, Denton ME, Costich DE, Jayasuriya CT, Saha M, Elshire R, Benscher D, Breseghello F, Munkvold J (2010) Cross-amplification of EST-derived markers among 16 grass species. Field Crops Res 118:28–35
Acknowledgements
Authors are thankful to Director, Sugarcane Research Institute, Shahjahanpur, India for conferring necessary accessories to perform this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest directly or indirectly and informed consent to publish this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, R.B., Singh, B. & Singh, R.K. Cross-taxon transferability of sugarcane expressed sequence tags derived microsatellite (EST-SSR) markers across the related cereal grasses. J. Plant Biochem. Biotechnol. 28, 176–188 (2019). https://doi.org/10.1007/s13562-019-00502-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-019-00502-6