Abstract
Lead poisoning in adults can affect the peripheral and central nervous systems, the kidneys, and blood pressure. Thus, the development of environment-friendly and simple methods for Pb2+ detection is of great importance. Herein, a label-free colorimetric method has been developed for the detection of Pb2+ based on the conformational switch from single-stranded DNA to G-quadruplex. The electrostatic interactions between DNA probe and gold nanorods (GNRs) induce GNRs to space closely. However, the electrostatic interaction is not strong enough to change the suspension state of GNRs. In the presence of Pb2+, the formation of G-quadruplexes increases the surface charge density around DNA, which is expected to strengthen the electrostatic interaction between the GNRs and the DNA. Therefore, the longitudinal absorption of GNRs decreased because the stronger interaction induced aggregation of GNRs. Importantly, the decrease in longitudinal absorption is proportional to concentration of Pb2+. By monitoring the change of absorbance, Pb2+ can be detected at a level of 3 nM with a linear range from 5 nM to 1 μM. The overall test only takes a few minutes and very little interference is observed from other metal ions. The major advantages of this method are its low cost, convenience, simplicity, sensitivity, and specificity.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Heavy metal pollution in the environment attracts increasing attention because it has severely adverse effect on human health. The contamination by heavy metal ions, particularly Pb2+, poses a serious threat to human health and environment. As lead is nondegradable, its persistence in the environment can produce toxic effects to plants and animals. Once introduced into the body, Pb2+ is a potential neurotoxin that can cause chronic inflammation of the kidney and heart, inhibit brain development, and decrease nerve conduction velocity [1–3]. The maximum level of lead in drinking water permitted by the US Environmental Protection Agency is 15 μg/L (∼72 nM) [4, 5]. These environmental and health problems of Pb2+ have prompted researchers to develop efficient methods for selective and sensitive assay of the heavy metal ion to understand its distribution and pollution potential. Therefore, development of a simple, sensitive, selective, economical, and practical method is highly demanded for environmental monitoring, food industry, and clinical diagnostics.
To monitor Pb2+ level, several methods have been developed, including inductively coupled plasma mass spectrometry [6, 7], atomic fluorescence spectrometry [8, 9], atomic absorption spectroscopy [10], reversed-phased high-performance liquid chromatography [11], and so on. Even with sensitivity and accuracy, there also share some disadvantages, such as time-consuming, expensive, and/or require sophisticated equipment, etc. To overcome these limitation and drawbacks, a variety of sensors have been developed to rapidly detect lead with high selectivity and sensitivity [12–26]. Among them, colorimetric sensors offer a promising approach for facile tracking of metal ions in biological, toxicological, and environmental samples. Lu and co-workers have developed a series of functional DNAzyme-based Pb2+ sensors by using GNPs. The detection range of the sensor could be tuned from 3 nM to 1 μM [19–25]. Dong and co-workers reported a DNAzyme-based colorimetric sensor for Pb2+; the detection limit was 32 nM [26]. In this paper, we aim to develop an ultrasensitive, environmentally friendly, yet simple method for colorimetric detection of Pb2+ ion.
Among the various nanostructures of gold, nanorods have attracted wide attention due to their versatile structures and special localized surface plasmon resonance [27, 28]. Gold nanorods (GNRs) have two directional electron oscillations in response to the polarization of the incident light. The absorption along the longer axis referred to as the longitudinal band is stronger (usually >600 nm) and the shorter axis one (around 520 nm) is called the transverse band. GNRs are normally passivated by positively charged surfactants, which give not only high stability but also positively charged surface. Gold nanorods as a typical anisotropic metal nanostructure possess many unique physical properties that have been widely applied in the field of medical imaging [29, 30] and biological sensors [31–39]. Mann et al. reported the specific organization of short GNRs into anisotropic three-dimensional aggregates by DNA hybridization [36]. He et al. utilized the optical and chemical properties of the GNRs designed a GNRs-quantum dots (QDs) quenching system for sensitive DNA detection [37]. Zhu et al. demonstrate a near infrared sensing system for the detection of human IgG based on the FRET between QDs and GNRs [38]. Ma et al. took advantage of the localized surface plasmon resonance properties of unmodified GNR detection DNA [39].
Herein, a label-free colorimetric method has been developed for the detection of Pb2+ in aqueous solution using gold nanorods as signal probe. By monitoring the change of longitudinal absorption of GNRs, the detection limit of Pb2+ was 3 nM. Circular dichroism (CD) and transmission electron microscope (TEM) measurements were adopted to further confirm these occurred interactions. Experimental results revealed that the developed method could be applied to monitor the existence of traces of Pb2+ ions in aqueous solution with high selectivity.
Experimental section
Chemicals
Unless otherwise indicated, all reagents and solvents were purchased in their highest available purity and used without further purification or treatment. HAuCl4·4H2O was purchased from Sigma (San Diego, USA). DNA (5′-GGGTGGGTGGGTGGGT-3′, DNA) was synthesized by Shanghai Sangon Biotechnology Co. (Shanghai, China) and used without further purification. The oligonucleotide stock solutions were prepared with a Tris–HAc buffer (pH 7.4) and kept frozen. Metal ion solutions were prepared from nitrate salts. Millipore Milli-Q (18.2 MΩ cm) water was used in all experiments.
Instrumentation
UV–Vis absorption spectra were recorded by using a Hitachi U-3900H UV–Vis Spectrophotometer (Tokyo, Japan). CD spectra were measured on a Chirascan Circular Dichroism Spectrometer (Applied Photophysics Ltd, London, England). The TEM images of GNRs were taken by using a JEM-2100 transmission electron microscope (Jeol Co. Ltd, Tokyo, Japan).
Synthesis of gold nanorods
Gold nanorods were synthesized by using a seed-mediated, surfactant-assisted growth method in a two-step procedure [31, 32]. Briefly, colloidal gold seeds were first prepared by mixing aqueous solutions of cetyltrimethylammonium bromide (CTAB, 0.1 M, 7.5 mL) and hydrogen tetrachloroaurate (III) hydrate (1 %, 0.098 mL). Freshly prepared aqueous solution of sodium borohydride (0.01 M, 0.6 mL) was then added. The colloidal gold seed solutions (0.215 mL) were then injected into an aqueous growth solution of CTAB (0.1 M, 47.6 mL), hydrogen tetrachloroaurate (III) hydrate (1 %, 0.788 mL), silver nitrate (0.01 M, 0.3 mL), and freshly prepared ascorbic acid (0.1 M, 0.32 mL). The nanorods were purified by several cycles of suspension in ultrapure water, followed by centrifugation. They were isolated in the precipitate, and excess CTAB was removed in the supernatant. Then, it was stored in a refrigerator at 4 ºC before being used. The nanorods were characterized by absorption spectroscopy.
UV–Vis measurements
Typically, the UV–Vis spectra of GNRs were recorded on UV–Vis spectrophotometer (Tokyo, Japan). Then, a certain quantity of GDNA was added into GNR suspension solution. After thorough mixing, a suitable Pb2+ was added to the mixture. The solution is vortexed thoroughly and used for the UV–Vis absorption spectra measurement experiment. The UV–Vis spectra of GNRs were recorded on a UV–Vis spectrophotometer with the wavelength range from 400 to 900 nm.
CD spectroscopy
The CD spectra of DNA oligonucleotides were measured for 2 μM DNA total strand concentration using a Chirascan Circular Dichroism Spectrometer (Applied Photophysics Ltd, London, England). CD spectra were recorded using a quartz cell of 1-mm optical path length and an instrument scanning speed of 100 nm/min with a response time of 2 s at room temperature. CD spectra were obtained by taking the average of three scans made from 200 to 350 nm. All DNA samples at a final concentration of 2 μM were dissolved in Tris–HCl buffer and heated to 90 °C for 5 min, gradually cooled to room temperature, and incubated at 4 °C overnight.
TEM
TEM images were obtained using a JEM-2100 transmission electron microscope (Jeol Co. Ltd, Tokyo, Japan). Samples were prepared on 400 mesh Cu grids coated with a thin layer of carbon (EM Sciences). The solution (5.00 μL) was pipetted onto the surface of the grid and allowed to dry in air. GNRs can grow out to 10 nm wide and up to 30 nm long with an aspect ratio of ∼3.
Results and discussion
Sensing mechanism
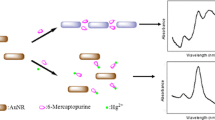
The design rationale is illustrated in Fig. 1. The assay is based on positively charged GNRs having a higher affinity to G-quadruplex DNA than ssDNA because the surface charge density of G-quadruplex DNA is much larger than that of ssDNA [40, 41]. In the absence of Pb2+, the electronic attraction between DNA and GNRs brought the DNA and GNRs into close proximity, which induced slight change of absorption spectrum of GNRs. However, upon the addition of Pb2+, electrostatic interactions obviously strengthened due to the formation of G-quadruplex, resulting in a decrease of longitudinal absorption of GNRs. Therefore, Pb2+ can be simply and directly detected by monitoring the change of absorbance or color.
It is known that the UV absorption spectra of GNRs display two bands assigned to the transversal and longitudinal modes of electronic oscillations, and the locations depend on the aspect ratio. As Fig. 2 shows, the two absorption bands for our prepared GNRs are located around 520 and 786 nm. The broadening of the transverse band of GNRs between 520 and 600 nm is ascribed to the presence of GNRs with different aspect ratios and gold nanoparticles with different sizes and shapes. When DNA was added to the GNR suspension, they attracted GNRs near because of the electrostatic interactions between the anionic backbone phosphates of oligonucleotides and the cationic surfactant bilayer around the nanorods. However, the electrostatic interaction is not strong enough to change the suspension state of GNRs. So, there is little change in the UV–Vis absorption spectra. Curve C in Fig. 2a shows the absorption spectrum of the GNRs in the presence of ssDNA upon addition of 5 nM Pb2+. Upon the addition of the Pb2+, the transverse absorption peak of the GNRs shows slight red shift, while the intensity of the longitudinal absorption shows a distinct decrease and a blueshift. This result suggests that GNRs assembled in the side-by-side mode due to the electrostatic interaction between the positive charge of GNRs and the negative charge of the phosphate backbone of the G-quadruplex DNA induced by Pb2+. It is consistent with the previous reports that the longitudinal band of GNRs is more sensitive to the local environment around GNRs than the transverse band [42–45]. This result suggested that the aggregation of GNRs takes place in a side-by-side manner [46, 47].
To further study the rationality and reliability of this assay, more control experiments have been performed. First, the influence of Pb2+ on the absorption spectrum of GNRs was studied. We investigated the absorption spectrum of GNRs in the presence of Pb2+ without ssDNA. As shown in Fig. 2b, the absorption spectrum of GNRs had no fundamental change without ssDNA as the concentration of Pb2+ increased. It is therefore obvious that the Pb2+ itself did not influence the longitudinal absorption of GNRs, but rather the decrease of absorbance in the presence of Pb2+ (Fig. 2a) is mainly caused by G-quadruplex formation which strengthened electrostatic interaction between GNRs and DNA. Then, the sensing mechanism was further studied by TEM images. From images a and b of Fig. 3, we found that GNRs are well dispersed in the aqueous medium in the absence and presence of DNA. However, GNRs obviously aggregated when Pb2+ was added into the GNRs/DNA solution in image C, which is accordance with the UV–Visible absorption measurements. Therefore, this colorimetric method was again demonstrated to be feasible and reasonable for the detection of Pb2+.
Identification of G-quadruplex formation
CD is a sensitive technology able to study the configuration inversion of DNA, which could report the structural variations intrinsically and kinetically [48]. To further confirm the formation of G-quadruplex, CD measurement is utilized to monitor the conformation change of DNA probe in the different cases. Figure 4a shows the CD spectra for the titration of the DNA with increasing amounts of Pb2+. The CD spectra of ssDNA at room temperature exhibited a positive band around 265 nm. Upon the addition of 2 μM Pb2+ to the ssDNA, a dramatic change in the CD spectrum was observed. The maximum at 265 nm was gradually increased. Meanwhile, a small positive peak appears near 320 nm, which indicated G-quadruplex formation [15, 49–52]. As the Pb2+ concentration increased to 5 μM, we observed a concentration-dependent enhancement of the positive peak around 265 and 320 nm and the negative peak around 240 nm. This CD spectrum suggests the coexistence of the parallel G4 structure with a small amount of the antiparallel one. As we know, K+ is highly able to stabilize the G4 structure. Therefore, the conformation change of DNA probe induced by K+ has also been studied. It is clear from Fig. 4b that the CD spectra of DNA probe slightly changed, and no new peak was observed around 320 nm. The DNA probe used in this work is named T30695. Wang et al. previously reported that the DNA melting experiments of K+-T30695 and Pb2+-T30695 have different stability under the same conditions. The stability of K+-T30695 is much lower than that of Pb2+-T30695 [15]. CD measurements revealed that the formation of G-quadruplex in the presence of Pb2+ led to the change of longitudinal band of GNRs.
CD spectra for characterizing the structural conversion of DNA (2 μM) in the absence and presence of Pb2+ (a) and K+ (b). a (a) without Pb2+; (b) 2 μM Pb2+; (c) 5 μM Pb2+. b (a) without K+; (b) 5 μM K+; (c) 10 μM K+; (d) 20 μM K+; (e) 50 μM K+. All solutions were prepared in 5 mM Tris–acetate (pH 7.4)
Selectivity and sensitivity
Selectivity was an important issue to estimate the performance of a sensor. So we tested the selectivity of the proposed method by comparing the absorbance changes of GNRs/DNA caused by Pb2+ and a variety of environmentally relevant metal ions, including Ag+, K+, Cd2+, Cu2+, Ni2+, Ba2+, Zn2+, Hg2+, Mg2+, Ca2+, Fe3+, and Al3+. Figure 5a illustrates the absorption intensity change (∆A = A 0 − A) where A 0 and A are the absorption intensity of GNRs/DNA in the absence and presence of different metal ions, respectively. It is obvious that all the metal ions except Pb2+ exhibited little variations in the extinction intensity, which is important and helpful in validation of the method to meet the selectivity requirements of the Pb2+ assay in environmental and biological fields. These results clearly reveal that our detection method has high selectivity against other interfering metal ions. As shown in Fig. 5b, the color of GNRs/DNA solution changed from red to blue when 2 μM Pb2+ was added, which indicated the aggregation of GNRs. However, other metal ions have no effect on the GNRs/DNA solution, even when the concentration of metal ions reached 5 μM. Therefore, this colorimetric method was again demonstrated to be feasible and reasonable for the selective detection of Pb2+.
Selectivity of the colorimetric method. a Absorbance change of GNRs/ssDNA in the presence of metal ions. The concentration of Pb 2+ is 2 μM, and the concentration of other metal ions is 5 μM. b Photographs of GNRs and GNRs/DNA in the absence and presence of metal ions. The concentration of Pb 2+ is 2 μM, and the concentration of other metal ions is 5 μM
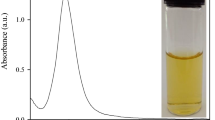
To evaluate the sensitivity of Pb2+ detection, the different concentrations of Pb2+ were added into GNRs/DNA solution, respectively. As expected, the decrease of absorbance of GNRs can quantitatively reflect the amount of lead ion added. From Fig. 6, we can see that a dramatic decrease in the absorbance was observed with the increasing of Pb2+ concentration. Curve A in Fig. 6 shows the UV–Vis spectra of GNRs/ssDNA. However, longitudinal absorption of GNRs showed a gradual decrease with the concentration of Pb2+ increasing from 5 nM to 3 μM. Upon the addition of Pb2+, the decline of GNRs absorption and the blueshift of the longitudinal band was obversed, which can be ascribed to the strengthened interaction between DNA and GNRs due to the formation of G-quadruplex in the presence of Pb2+ because G-quadruplex has higher charge density than the ssDNA [53, 54]. As shown in the inset of Fig. 6, the proposed method exhibited a good linear response (R = 0.9932) of absorbance change against the logarithm of Pb2+ ion concentration over the range from 5 nM to 1 μM, indicating that Pb2+ can be sensitively detected by this method. The detection limit of Pb2+ was 3 nM, which was much lower than the EPA standard for the maximum allowable level 15 μg/L (72 nM) in drinking water.
Analysis of water
To demonstrate the application potential of our proposed method in environmental analysis, we applied it to analyze real tap water samples. The tap water sample was collected after discharging tap water for ∼20 min. Standard addition method was used to valuate the practicality of developed approach. All the water samples were spiked with Pb2+ at different concentration levels. The Pb2+ concentrations were calculated using standard curves prepared within the same day by our new approach. From Table 1, we can conclude that it is feasible for Pb2+ detection in real tap water samples.
Conclusion
In summary, a simple, direct, and cost-effective method has been developed for rapid detection of Pb2+ by using GNRs as colorimetric probe. The experimental results show that Pb2+ can be detected quickly and accurately with high sensitivity and selectivity against other heavy metal ions. Under the optimal conditions, this method was highly sensitive (LOD = 3 nM) and selective toward Pb2+ ions, with a linear detection range from 5 nM to 1 μM. From the summary in Table 1 (supporting information), we can conclude that the major advantages of this method are its simplicity, selectivity, and high sensitivity. It is of great theoretical and practical importance for the detection of heavy metal ions.
References
Laterra J, Bressler JP, Indurti RR, Belloni-Olivi L, Goldstein GW (1992) Proc Natl Acad Sci USA 89:10748–10752
Kim HN, Ren WX, Kim JS, Yoon JY (2012) Chem Soc Rev 41:3210–3244
Zocche JJ, Leffa DD, Damiani AP, Carvalho F, Mendonca RA, Santos CE, Boufleur LA, Dias JF, Andrade VM (2010) Environ Res 110:684–691
Huang KW, Cheng-Ju YCJ, Tseng WL (2010) Biosens Bioelectron 25:984–989
Chen YY, Chang HT, Shiang YC, Hung YL, Chiang CK, Huang CC (2009) Anal Chem 81:9433–9439
Liu HW, Jiang SJ, Liu SH (1999) Spectrochim Acta Part B 54:1367–1375
Bowins RJ, Mcnutt RH (1994) J Anal At Spectrom 9:1233–1236
Wagner EP, Smith BW, Winefordner JD (1996) Anal Chem 68:3199–3203
Neuhause RE, Panne U, Niessner R, Petrucci GA, Cavalli P, Omenetto N (1997) Anal Chim Acta 346:37–48
Weidenhamer JD (2007) J Chem Educ 84:1165–1166
Saito S, Danzaka N, Hoshi S (2006) J Chromatogr A 1104:140–144
Wang L, Jin Y, Deng J, Chen GZ (2011) Analyst 136:5169–5174
Song PS, Xiang Y, Xing H, Zhou ZJ, Lu Y (2012) Anal Chem 84:2916–2922
Zhao XH, Kong RM, Zhang XB, Meng HM, Liu WN, Tan WH, Shen GL, Yu RQ (2011) Anal Chem 83:5062–5066
Li T, Dong SJ, Wang EK (2010) J Am Chem Soc 132:13156–13157
Bui MPN, Li CA, Han KN, Pham XH, Seong GH (2012) Analyst 137:1888–1894
Fu XB, Qu F, Li NB, Luo HQ (2012) Analyst 137:1097–1099
Lee YF, Deng TW, Chiu WJ, Wei TY, Roy P, Huang CC (2012) Analyst 137:1800–1806
Liu JW, Lu Y (2003) J Am Chem Soc 125:6642–6643
Liu JW, Lu Y (2004) Chem Mater 16:3231–3238
Liu JW, Lu Y (2004) J Am Chem Soc 126:12298–12305
Liu JW, Lu Y (2004) Anal Chem 76:1627–1632
Liu JW, Lu Y (2005) J Am Chem Soc 127:12677–12683
Wang ZD, Lee JH, Lu Y (2008) Adv Mater 20:3263–3267
Mazumdar D, Liu JW, Lu G, Zhou JZ, Lu Y (2010) Chem Commun 46:1416–1418
Li T, Wang EK, Dong SJ (2010) Anal Chem 82:1515–1520
Juste JP, Santos IP, Liz-Marzán LM, Mulvaney P (2005) Coord Chem Rev 249:1870–1901
Keul HA, Möller A, Bockstaller MR (2007) Langmuir 23:10307–10315
Maltzahn GV, Centrone A, Park JH, Ramanathan R, Sailor MJ, Hatton TA, Bhatia SN (2009) Adv Mater 21:3175–3180
Wang HF, Huff TB, Zweifel DA, He W, Low PS, Wei A, Cheng JX (2005) Proc Natl Acad Sci USA 102:15752–15756
Huang XH, Neretina S, El-Sayed MA (2009) Adv Mater 21:4880–4910
Paraba HJ, Jung C, Lee JH, Park HG (2010) Biosens Bioelectron 26:667–673
York J, Spetzler D, Xiong FS, Frasch WDD (2008) Lab Chip 8:415–419
Li CZ, Male KB, Hrapovic S, Luong JHT (2005) Chem Commun 31:3924–3926
Sim HR, Wark AW, Lee HJ (2010) Analyst 135:2528–2532
Dujardin E, Hsin LB, Wang CRC, Mann S (2001) Chem Commun 14:1264–1265
Li X, Qian J, Jiang L, He SL (2009) Appl Phys Lett 94:063111
Liang GX, Pan HC, Li Y, Jiang LP, Zhang JR, Zhu JJ (2009) Biosens Bioelectron 24:3693–3697
Ma ZF, Tian L, Wang TT, Wang CG (2010) Anal Chim Acta 673:179–184
Jin Y, Chen GZ, Wang YX (2011) Gold Bull 44:163–169
Gou XC, Liu J, Zhang HL (2010) Anal Chim Acta 668:208–214
Pan BF, Cui DX, Ozkan C, Xu P, Huang T, Li Q, Chen H, Liu FT, Gao F, He R (2007) J Phys Chem C 111:12572–12576
Chang JY, Wu HM, Chen H, Ling YC, Tan WH (2005) Chem Commun 8:1092–1094
Nehl CL, Hafne JH (2008) J Mater Chem 18:2415–2419
Sudeep PK, Joseph ST, Thomas KG (2005) J Am Chem Soc 127:6516–6517
Sun ZH, Ni WH, Yang Z, Kou XS, Li L, Wang JF (2008) Small 4:1287–1292
Jain PK, Eustis S, El-Sayed MA (2006) J Phys Chem B 110:18243–18253
Paramasivan S, Rujan I, Bolton PH (2007) Methods 43:324–331
Li CL, Liu KT, Lin YW, Chang HT (2011) Anal Chem 83:225–230
Jing NJ, Rando RF, Pommier Y, Hogan ME (1997) Biochemistry 36:12498–12505
Smirnov I, Shafer RH (2000) J Mol Biol 296:1–5
Li T, Wang EK, Dong SJ (2009) J Am Chem Soc 131:15082–15083
Chen GZ, Jin Y, Wang L, Deng J, Zhang CX (2011) Chem Commun 47:12500–12502
He W, Huang CZ, Li YF, Xie JP, Yang RG, Zhou PF, Wang J (2008) Anal Chem 80:8424–8430
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (21075079), Program for New Century Excellent Talents in University (NCET-10-0557), and the Program for Changjiang Scholars and Innovative Research Team in 403 University (IRT 404 1070).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 73 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chen, G., Jin, Y., Wang, W. et al. Colorimetric assay of lead using unmodified gold nanorods. Gold Bull 45, 137–143 (2012). https://doi.org/10.1007/s13404-012-0057-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13404-012-0057-6