Abstract
Persian leopards Panthera pardus saxicolor have been extirpated from over 84% of their historic range and are now limited to rugged landscapes of West Asia and the Caucasus. Understanding and maintaining genetic diversity and population connectivity is important for preventing inbreeding and genetic drift, both of which can threaten population viability. All previous analyses of intraspecific genetic variation of West Asian leopards based on the NADH dehydrogenase subunit 5 gene have reported low mitogenomic diversity. In the current study, we sequenced 959 bp of the mtDNA cytochrome b gene to describe the spatial genetic structure of 22 wild Persian leopards across Iran, which hosts most of the subspecies extant range. The findings based on phylogenetic trees and median-joining network indicated that leopards from Iran formed a distinct subclade, i.e., P. p. saxicolor. The AMOVA analysis showed significant differentiation (88.55%) between the subclades of Persian leopards and other Asian leopards. The lowest levels of haplotype (0.247) and nucleotide (0.00078) diversity were estimated in Persian leopards from Iran. Mitochondrial genome sequencing revealed only two closely related haplotypes. There was no evidence for recent sudden demographic expansion scenario in Persian leopards. The low diversity in cytochrome b gene could potentially be brought about by selective pressure on mitochondria to adapt to oxidative stress and higher metabolic rates in cold environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a large-bodied, wide-ranging felid, the leopard Panthera pardus has evolved to persist across a diverse range of habitat types (Jacobson et al. 2016). Adapting to extreme heterogeneity across its vast global range has resulted in remarkable morphological variation, encouraging taxonomists to describe up to 27 putative subspecies (Ellerman and Morrison-Scott 1966; Herrington 1986), recently revised to only 8 subspecies (Kitchener et al. 2017). Leopards also show higher genetic diversity than many other big cats such tigers P. tigris (Luo et al. 2004), jaguars P. onca (Wultsch et al. 2016) and snow leopards P. unica (Janecka et al. 2017). Nonetheless, their heterozygosity is not spatially homogenous across their global range (Uphyrkina et al. 2001).
West Asian leopards have been the subject of controversies surrounding their taxonomy, phylogeography, population genetic structure, and variability (Khorozyan et al. 2006; Kitchener et al. 2017). A total of seven putative subspecies has been reported for the region (Herrington 1986), although many of them were recently identified as synonyms based on molecular analyses (Uphyrkina et al. 2001; Rozhnov et al. 2011; Farhadinia et al. 2015). Currently, two subspecies P. p. saxicolor = P. p. dathei = P. p. sindica = P. p. ciscaucasica = P. p. tulliana and P. p. nimr = P. p. jarvisi are proposed for West Asia (Kitchener et al. 2017). Several recent studies have provided understanding of taxonomy, phylogeography, spatial population structuring, and genetic variability based on ancient samples (Paijmans et al. 2018) as well as recent mitogenome sequencing in West Asian leopards, such as Iran (Farhadinia et al. 2015), the Caucasus (Rozhnov et al. 2011), and Pakistan (Asad et al. 2019).
Nonetheless, all previous analyses of intraspecific genetic variation of West Asian leopards have been limited to only one mitochondrial gene, the NADH dehydrogenase subunit 5 (NADH-5), which exhibits relatively high rates of mutation in leopards (Miththapala et al. 1996; Uphyrkina et al. 2001), accounting for 44 of 50 variable sites found in mtDNA regions of the leopards (Uphyrkina et al. 2001). Surprisingly, low mitogenomic diversity was reported in all studied populations of Persian leopards, from Pakistan to Iran and the Caucasus (Rozhnov et al. 2011; Farhadinia et al. 2015; Asad et al. 2019). Mitochondrial analyses of nearly 100 wild-born leopards revealed no more than 8 haplotypes across West Asia.
Mitochondrial markers have been extensively used to reveal the stochastic processes of coalescence and lineage sorting of leopards (Uphyrkina et al. 2001, 2002; Rozhnov et al. 2011; Farhadinia et al. 2015; Ropiquet et al. 2015; Paijmans et al. 2018). The objective of our study was to clarify if the low mitgenomic variation in Persian leopards is only seen on NADH-5 gene or other genes, with high variability in other leopard populations, will also show similar patterns in Persian leopards. Accordingly, the present study aims to evaluate the genetic variability of leopards using another mtDNA gene which has never been tested for West Asian populations. Within the leopard mitogenome, cytochrome b (cyt b) coding genes exhibit high levels of both inter- and intrapopulation variability (Uphyrkina et al. 2001; Ropiquet et al. 2015). We use a spatially representative sample of Persian leopards in Iran, which hosts 86% of the subspecies extant range (Jacobson et al. 2016). With the demand for leopard capturing and translocation in order to resolve the threat of severe livestock raiding (Weise et al. 2015; Farhadinia et al. 2018) or to restore the historic range of the species (Rozhnov et al. 2011; Breitenmoser et al. 2014), our findings are helpful for understanding spatial genetic structure and phylogeography of leopards in West Asia.
Materials and methods
Sample collection and DNA extraction
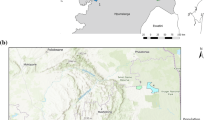
We obtained 22 tissue samples, from both muscle and skin, from wild-born dead leopards confiscated from poachers or live individuals captured for a satellite telemetry program, all with known geographic origin in Iran (Fig. 1 and Supplementary Table 1; permissions number 31/12630, 93/16270, 93/16258, and 93/16270 issued by the Iranian Department of Environment). Samples were collected from three major regions of leopard range associated with major mountain chains, namely, Zagros, Alborz, and Kopet Dag (Fig. 1). All samples were stored in 96% ethanol until further analysis. Total DNA was extracted from leopard tissue samples using a standard phenol/chloroform method described in Sambrook et al. (1989), employing negative controls for each batch of extractions. No detectable DNA (as visualized on agarose gel) or polymerase chain reaction (PCR) product was obtained from any of the negative controls.
Spatial distribution of sampling locations and cyt b haplotypes of Persian leopard in Iran. Leopard extant range is obtained from Jacobson et al. (2016)
DNA sequencing
We used leoF: GACYAATGATATGAAAAACCATCGTTG/Leo-R: GTTCTCCTTTTTTGGTTTACAAGAC for a fragment of 959 bp cyt b amplification (Ropiquet et al. 2015). For each reaction, 35 cycles were performed with 5 min initial denaturation at 95 °C followed by denaturation at 94 °C for 0.5 min, 1.5 min annealing at 50 °C, 1 min extension at 72 °C, followed by a 5 min final extension at 72 °C. The PCR products were checked in 1.5% agarose gels. Purification was carried out using column-based purification kits (Millipore) using a vacuum for filtering. Sequences were visualized using an ABI-3730xl genetic analyzer (Applied Biosystems; http://www.appliedbiosystems.com).
Data analysis
We aligned sequences using the progressive pairwise alignment implemented in Geneious® 11.0.3 to a 959 bp segment of mtDNA cyt b. New sequences were combined with 50 GenBank entries (Supplementary Table 2) to create the data set in order to clarify the phylogenetic relationships among leopard (P. pardus) populations. Lion (P. leo) (KC701376 and KM374704) and tiger (P. tigris) (AF053053, AF053054, and DQ151551) were used as outgroups, following Ropiquet et al. (2015).
Substitution saturation was analyzed using DAMBE6 (Xia 2017). The best-fit substitution models were evaluated using the corrected Akaike Information Criterion (AICc) and the Bayesian information criterion (BIC) calculated in PartitionFinder 2.1.1 (Lanfear et al. 2017). The Bayesian inference was reconstructed using MrBayes 3.2 (Ronquist et al. 2012) in four Monte Carlo Markov chains for 50,000,000 generations and tree sampling every 100 generations. IQ-TREE 1.6 (Nguyen et al. 2015) was used for inferring the maximum likelihood tree with 10,000 replicates. The median-joining network was inferred using Network 10.0 (available at http://www.fluxusengineering.com/sharenet.htm), in order to reconstruct the relationships among haplotypes.
AMOVA (analysis of molecular variance), ΦST (genetic distance), and Nm (gene flow) statistics were calculated with 10,000 iterations using Arlequin 3.5 (Excoffier and Lischer 2010). Haplotype diversity, nucleotide diversity, number of polymorphic sites, number of haplotypes, and average number of nucleotide differences were calculated using DnaSp 5 (Librado and Rozas 2009). The neutrality tests of Tajima’s D (Tajima 1989), Fu’s FS (Fu 1997), and R2 (Ramos-Onsins and Rozas 2002) were calculated in Arlequin 3.5 (Excoffier and Lischer 2010) and DnaSp 5 (Librado and Rozas 2009) to test the hypothesis of selective neutrality and to examine for evidence of a recent demographic expansion (Ramírez-Soriano et al. 2008).
Results
Totally, 22 samples of Persian leopard across Iran were sequenced for the 959 bp segment of the mtDNA cyt b. A total of two haplotypes were detected within the 22 individuals from Iran, namely, IRAN4 (N = 21 individuals) and IRAN5 (N = 3 individuals), which have been deposited in GenBank with accession numbers MK028952 and MK028953. The nucleotide differences between the two haplotypes were three transitions.
Haplotype IRAN4 occurred commonly across the study area, whereas the haplotype IRAN5 was detected only in the Alborz region (Fig. 1). Additional sequences from global geographic range of the species extracted from GenBank were used in the phylogenetic analyses, where the final dataset contained 72 sequences and 22 haplotypes (Table 3). Result of substitution saturation indicated that base substitutions did not reach saturation, so the dataset can be used in phylogenetic analyses.
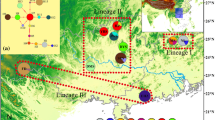
The phylogenetic analyses (Bayesian and maximum likelihood trees) resulted in identical topologies included two clades: Asian and African. Each of these clades was subdivided into two subclades (Fig. 2). These clades and subclades are very well supported by bootstrap values and posterior probabilities. The leopard living in Iran formed a distinct subclade that fell into the Asian clade. The median-joining network (Fig. 3) also demonstrated the subclades and clades detected in the phylogenetic trees.
Phylogenetic relationships of Persian leopards (Asian subclade A) with other identified subclades of leopard (P. pardus) derived from a sequence of 959 bp mtDNA cyt b and rooted with P. leo and P. tigris. The numbers on the branches are bootstrap values and posterior probabilities in the maximum likelihood and Bayesian analysis, respectively. IRAN4 and IRAN5 denote detected haplotypes in the present study. For detailed information on sequences, see Table S1
The pairwise ΦST values indicated significant differences among the identified subclades (ranging from 0.732 to 0.945). The lowest distance (0.732) occurred between two African subclades, with Nm value of 0.183. The highest distance (0.945) estimated between the Asian subclade A (Persian subclade) and African subclade A, with Nm value of 0.029 (Table 1). Persian leopards exhibited smaller distance to other Asian leopards (India and East Asia) than African leopards. The AMOVA results revealed significant differentiations between the two identified subclades within each of the Asian (genetic variations between subclades = 88.55%) and African (genetic variations between subclades = 73.17%) clades (Table 2).
The lowest levels of haplotype (0.247) and nucleotide (0.00078) diversity were estimated in Persian leopards (Asian subclade A), whereas the highest values of haplotype (0.942) and nucleotide (0.00795) diversity were calculated in the African subclades (Table 3). The results of the neutrality tests (Fu’s FS = 2.272, Tajima’s D = − 0.260, R2 = 0.1234) were not significant for Persian leopards, where negative value was only calculated for Tajima’s D test. Thus, the sudden population expansion scenario can be rejected in Iran. However, the neutrality tests for the African subclade A were significantly negative (Fu’s FS = − 5.873, p < 0.01; Tajima’s D = − 1.21; R2 = 0.835, p < 0.05).
Discussion
We found low mitogenome diversity for Persian leopards in Iran without substantial population subdivision, based on the mtDNA cyt b gene. Our data supported a common maternal genealogy for leopards in West Asia and the Caucasus (Rozhnov et al. 2011; Farhadinia et al. 2015) without evidence of demographic expansion. The unstructured matrilines suggest that Iran’s leopards currently form a single gene pool, although nuclear markers are needed to confirm this hypothesis.
Previous studies found low haplotype diversity in leopards living in West Asian mountains, such as Pakistan, Iran, Turkmenistan, and the Caucasus based on NADH-5 gene (Rozhnov et al. 2011; Farhadinia et al. 2015; Asad et al. 2019). Our analyses using another mitochondrial marker, i.e., cyt b confirmed the low variability in mitogenomic sequences of leopards. Similarly, snow leopards, another Panthera species roaming Asia’s rugged mountains (Janecka et al. 2017), show low haplotype diversity. In contrast, other wide-ranging large carnivores in Asian highlands such as brown bear Ursus arctos and Himalayan gray wolf Canis himalayensis show remarkable haplotype diversity (Murtskhvaladze et al. 2010; Ashrafzadeh et al. 2016; Werhahn et al. 2018). Felids, especially Panthera species, have the most recent divergence time and consequently a lower molecular evolution rate compared with Ursids and Canids (Wayne et al. 1991). This has resulted in little intraspecific genetic diversity and shorter inter-specific genetic distances (Slattery et al. 2000; Kim et al. 2016).
The mitochondrial genomes of leopards are highly variable across most of their global range (Uphyrkina et al. 2001; Ropiquet et al. 2015; Anco et al. 2017). There are two potential hypotheses for low haplotype diversity in Persian leopards. From the demographic perspective, this scenario can be inherited from bottleneck events in recent or historic times (Ellegren and Galtier 2016), similar to Amur tiger P. t. altaica (Russello et al. 2004), Amur leopard P. p. orientalis (Uphyrkina et al. 2002), and Asiatic cheetah Acinonyx jubatus venaticus (Charruau et al. 2011). All these have experienced a recent or ongoing demographic bottleneck with a small founding population. Although Persian leopards survived widespread eradication attempts in the Caucasus during the 1900s, leaving a few dozen (Khorozyan and Abramov 2007), no contemporary demographic bottleneck is known from Iran, with an estimated population of between 550 and 850 individuals and wide distribution by the end of the 1900s (Kiabi et al. 2002) with some areas harboring high leopard densities (Farhadinia et al. 2019). It is therefore less likely that demography can comprehensively explain the current genetic pattern in Persian leopards.
Alternatively, it might be associated with the selective pressure on endothermic vertebrates to cope with the combined effects of hypoxia and cold stresses at high altitudes (Cheviron and Brumfield 2012). There, they need adaptive evolution of the mitochondrial oxidative phosphorylation system to regulate oxygen usage and energy metabolism (da Fonseca et al. 2008; Hassanin et al. 2009; Werhahn et al. 2018). Similarly for another montane-living large felid, Janecka et al. (2017) suggested that the snow leopard mitogenome may have undergone selective pressure in hypoxic environments, resulting in low mitochondrial diversity. Future researches can test this hypothesis using leopards sampled at different elevations analyzed for candidate hypoxia genes (Zhang et al. 2014; Werhahn et al. 2018).
These findings support a distinction between Asian and African leopard populations. We found additional strong support for the divergence between Persian leopards and other Asian leopards. In parallel with previous works (Uphyrkina et al. 2001; Rozhnov et al. 2011; Farhadinia et al. 2015), our study confirms a monophyletic group for the Iranian female lineage. The pattern can be expanded to leopards from other West Asian countries within the range of Persian leopard to share similar monophyletic group with Iran (Rozhnov et al. 2011; Asad et al. 2019). Lack of substantial population subdivision suggests that Iran’s leopards currently form a single gene pool.
We acknowledge that our results are limited by the use of mtDNA, and consequently single locus data. However, our spatially representative samples from the majority of Persian leopard range along with previous sequencing from other range countries in West Asia will provide the basic understanding in mitochondrial variability, partitioning, and phylogeographic patterns of leopards in West Asia. Nonetheless, further studies are encouraged to apply multi-locus sampling to reveal fine-scale population structure and variability. Also, leopard samples from central Asian countries such as Afghanistan, Turkmenistan, and Kazakhstan can further enhance our understanding about the phylogeorgraphy and taxonomy of Persian leopards. Importantly, in most countries in West Asia and the Caucasus, the majority of the remaining leopard range is along international borders, and thus in most countries, conservation of these subspecies is dependent on transboundary collaboration (Farhadinia et al. 2020). Therefore, a trans-national genetic analysis with samples representing the range of the P. p. saxicolor to understand the gene flow and spatial differentiation is desirable.
References
Anco C, Kolokotronis S-O, Henschel P, et al (2017) Historical mitochondrial diversity in African leopards (Panthera pardus) revealed by archival museum specimens. Mitochondrial DNA Part A 1–19. https://doi.org/10.1080/24701394.2017.1307973
Asad M, Martoni F, Ross JG, Waseem M, Abbas FI, Paterson AM (2019) Assessing subspecies status of leopards (Panthera pardus) of northern Pakistan using mitochondrial DNA. PeerJ 7:e7243
Ashrafzadeh MR, Kaboli M, Naghavi MR (2016) Mitochondrial DNA analysis of Iranian brown bears (Ursus arctos) reveals new phylogeographic lineage. Mamm Biol für Säugetierkd 81:1–9
Breitenmoser U, Pittet M, Avgan B et al (2014) Recovery of the leopard in Azerbaijan. Zoological Society of London, United Kingdom
Charruau P, Fernandes C, OROZCO-terWENGEL P et al (2011) Phylogeography, genetic structure and population divergence time of cheetahs in Africa and Asia: evidence for long-term geographic isolates. Mol Ecol 20:706–724
Cheviron ZA, Brumfield RT (2012) Genomic insights into adaptation to high-altitude environments. Heredity (Edinb) 108:354–361
da Fonseca RR, Johnson WE, O’Brien SJ et al (2008) The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics 9:119
Ellegren H, Galtier N (2016) Determinants of genetic diversity. Nat Rev Genet 17:422–433
Ellerman JR, Morrison-Scott TCS (1966) Family Felidae. In: Checklist of Palaearctic and Indian Mammals 1758 to 1946. Trustees of the British museum (natural history), London, pp 300–321
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Farhadinia MS, Farahmand H, Gavashelishvili A, Kaboli M, Karami M, Khalili B, Montazamy S (2015) Molecular and craniological analysis of leopard, Panthera pardus (Carnivora: Felidae) in Iran: support for a monophyletic clade in Western Asia. Biol J Linn Soc 114:721–736
Farhadinia MS, Johnson PJ, Hunter LTB, Macdonald DW (2018) Persian leopard predation patterns and kill rates in the Iran–Turkmenistan borderland. J Mammal 99:713–723
Farhadinia MS, McClintock BT, Johnson PJ et al (2019) A paradox of local abundance amidst regional rarity: the value of montane refugia for Persian leopard conservation. Sci Rep 9:14622
Farhadinia MS, Rostro-García S, Leng L, Kamler JF, Spalton A, Shevtsova E, Khorozyan I, AL-Duais M, Ge J, Macdonald DW (2020) Big cats in borderlands: challenges and implications for transboundary conservation of Asian leopards. Oryx 54 (in press)
Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Hassanin A, Ropiquet A, Couloux A, Cruaud C (2009) Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). J Mol Evol 68:293–310
Herrington SJ (1986) Phylogenetic relationships of the wild cats of the world. University of Kansas, Systematics and Ecology
Jacobson AP, Gerngross P, Lemeris JR et al (2016) Leopard (Panthera pardus) status, distribution, and the research efforts across its range. PeerJ 4:e1974
Janecka JE, Zhang Y, Li D, Munkhtsog B, Bayaraa M, Galsandorj N, Wangchuk TR, Karmacharya D, Li J, Lu Z, Uulu KZ, Gaur A, Kumar S, Kumar K, Hussain S, Muhammad G, Jevit M, Hacker C, Burger P, Wultsch C, Janecka MJ, Helgen K, Murphy WJ, Jackson R (2017) Range-wide snow leopard phylogeography supports three subspecies. J Hered 108:597–607
Khorozyan I, Abramov AV (2007) The leopard Panthera pardus (Carnivora: Felidae) and its resilience to human pressure in the Caucasus. Zool Middle East 41:11–24
Khorozyan I, Gennady F, Baryshnikov GF, Abramov AV (2006) Taxonomic status of the leopard, Panthera pardus (Carnivora, Felidae) in the Caucasus and adjacent areas. Russ JTheriol 5:41–52
Kiabi B, Dareshouri B, Ghaemi R, Jahanshahi M (2002) Population status of the Persian leopard (Panthera pardus saxicolor Podock, 1927) in Iran. Zool Middle East 26:41–47
Kim S, Cho YS, Kim H-M, Chung O, Kim H, Jho S, Seomun H, Kim J, Bang WY, Kim C, An J, Bae CH, Bhak Y, Jeon S, Yoon H, Kim Y, Jun JH, Lee HJ, Cho S, Uphyrkina O, Kostyria A, Goodrich J, Miquelle D, Roelke M, Lewis J, Yurchenko A, Bankevich A, Cho J, Lee S, Edwards JS, Weber JA, Cook J, Kim S, Lee H, Manica A, Lee I, O’Brien SJ, Bhak J, Yeo JH (2016) Comparison of carnivore, omnivore, and herbivore mammalian genomes with a new leopard assembly. Genome Biol 17:211. https://doi.org/10.1186/s13059-016-1071-4
Kitchener AC, Breitenmoser-Würsten C, Eizirik E, et al (2017) A revised taxonomy of the Felidae: the final report of the cat classification task force of the IUCN cat specialist group. Cat News Spec Issue 1–80
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34:772–773
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Luo SJ, Kim JH, Johnson WE et al (2004) Phylogeography and genetic ancestry of tigers (Panthera tigris). PLoS Biol 2:2275–2293
Miththapala S, Seidensticker J, O’Brien SJ (1996) Phylogeographic subspecies recognition in leopards (Panthera pardus): molecular genetic variation. Conserv Biol 10:1115–1132
Murtskhvaladze M, Gavashelishvili A, Tarkhnishvili D (2010) Geographic and genetic boundaries of brown bear (Ursus arctos) population in the Caucasus. Mol Ecol 19:1829–1841
Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Paijmans JLA, Barlow A, Foerster DW et al (2018) Historical biogeography of the leopard (Panthera pardus) and its extinct Eurasian populations. BMC Evol Biol 18:156
Ramírez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A (2008) Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics 179:555–567
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Ronquist F, Teslenko M, Van Der Mark P et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Ropiquet A, Knight AT, Born C, Martins Q, Balme G, Kirkendall L, Hunter L, Senekal C, Matthee CA (2015) Implications of spatial genetic patterns for conserving African leopards. C R Biol 338:728–737
Rozhnov VV, Lukarevskiy VS, Sorokin PA (2011) Application of molecular genetic characteristics for reintroduction of the leopard (Panthera pardus) in the Caucasus. Dokl Biol Sci 437:280–285
Russello MA, Gladyshev E, Miquelle D, Caccone A (2004) Potential genetic consequences of a recent bottleneck in the Amur tiger of the Russian far east. Conserv Genet 5:707–713
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. edition. New York Cold Spring Harb Lab
Slattery JP, Murphy WJ, O’Brien SJ (2000) Patterns of diversity among SINE elements isolated from three Y-chromosome genes in carnivores. Mol Biol Evol 17:825–829
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Uphyrkina O, Johnson WE, Quigley HB, Miquelle D, Marker L, Bush M, O'Brien SJ (2001) Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Mol Ecol 10:2617–2633
Uphyrkina O, Miquelle DG, Quigley HB, et al (2002) Conservation genetics of the far eastern leopard (Panthera pardus orientalis) 93:303–311
Wayne RK, van Valkenburgh B, O’Brien SJ (1991) Molecular distance and divergence time in carnivores and primates. Mol Biol Evol 8:297–319
Weise FJ, Lemeris J Jr, Stratford KJ, van Vuuren RJ, Munro SJ, Crawford SJ, Marker LL, Stein AB (2015) A home away from home: insights from successful leopard (Panthera pardus) translocations. Biodivers Conserv 24:1755–1774
Werhahn G, Senn H, Ghazali M, Karmacharya D, Sherchan AM, Joshi J, Kusi N, López-Bao JV, Rosen T, Kachel S, Sillero-Zubiri C, Macdonald DW (2018) The unique genetic adaptation of the Himalayan wolf to high-altitudes and consequences for conservation. Glob Ecol Conserv 16:e00455
Wultsch C, Caragiulo A, Dias-Freedman I, Quigley H, Rabinowitz S, Amato G (2016) Genetic diversity and population structure of Mesoamerican jaguars (Panthera onca): implications for conservation and management. PLoS One 11:e0162377
Xia X (2017) DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. J Hered 108:431–437
Zhang W, Fan Z, Han E, Hou R, Zhang L, Galaverni M, Huang J, Liu H, Silva P, Li P, Pollinger JP, du L, Zhang XY, Yue B, Wayne RK, Zhang Z (2014) Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet plateau. PLoS Genet 10:e1004466
Acknowledgments
We are grateful to various provincial offices of the DoE and private collections who donated samples. K. Khosraviani and N. Ghovati assisted in lab experiments.
Availability of data and material
All sequences have been deposited in GenBank with accession numbers MK028952 and MK028953.
Code availability
All software are cited in the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
For dead samples; no ethical approval was needed. Sampling was conducted based on permissions number 31/12630 and 93/16258 issued by the Iranian Department of Environment. We also obtained samples from live individuals captured for a satellite telemetry study in Tandroueh National Park. The Iranian Department of Environment reviewed all sampling, trapping, and handling procedures and approved permits for the work conducted (93/16270). The trapping and handling protocol was also approved by the University of Oxford’s Ethical Review Committee (BMS-ERC-160614).
Consent to participate
All authors agreed to participate in this study and co-authorship.
Consent for publication
All authors agreed with the content and that all gave explicit consent to submit.
Additional information
Communicated by: Jan M. Wójcik
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 48.6 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Farhadinia, M.S., Ashrafzadeh, M.R., Senn, H. et al. Is there low maternal genetic variation in West Asian populations of leopard?. Mamm Res 65, 701–708 (2020). https://doi.org/10.1007/s13364-020-00510-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13364-020-00510-1