Abstract
Significant effort is being employed to utilize the inherent speed and sensitivity of mass spectrometry for rapid structural determination of proteins; however, a thorough understanding of factors influencing the transition from solution to gas phase is critical for correct interpretation of the results from such experiments. It was previously shown that combined use of action excitation energy transfer (EET) and simulated annealing can reveal detailed structural information about gaseous peptide ions. Herein, we utilize this method to study microsolvation of charged groups by retention of 18-crown-6 (18C6) in the gas phase. In the case of GTP (CEGNVRVSRE LAGHTGY), solvation of the 2+ charge state leads to reduced EET, whereas the opposite result is obtained for the 3+ ion. For the mini-protein C-Trpcage, solvation by 18C6 leads to dramatic increase in EET for the 3+ ion. Examination of structural details probed by molecular dynamics calculations illustrate that solvation by 18C6 alleviates the tendency of charged side chains to seek intramolecular solvation, potentially preserving native-like structures in the gas phase. These results suggest that microsolvation may be an important tool for facilitating examination of native-like protein structures in gas phase experiments.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proteins play critical roles in the biochemistry of life. The functionality of proteins is closely connected to their three-dimensional structure, which is therefore an important target for research. Within a cell, the solvent is aqueous and highly buffered. In addition, a plethora of other biomolecules are present in high abundance (i.e., the cellular matrix is ‘crowded’). Interactions with solvent and other biomolecules influence protein structure; therefore, it is desirable to retain such elements during structural characterization. Unfortunately, this goal is never achieved with complete fidelity, and compromises must be made regardless of the experimental technique to simplify the system for analysis.
Presently, there is significant interest in developing mass spectrometry (MS) based methods for examining protein structure in the gas phase because of inherent advantages such as speed and sensitivity [1, 2]. For example, ion mobility (IM) can be coupled with MS to obtain collision cross sections, yielding information about overall molecular shape [3–8]. Collision induced unfolding has recently proven to be valuable in distinguishing antibody isoforms by comparing “fingerprint plots” generated by differences in unfolding of the isoforms in an ion trap prior to IM-MS analysis [9]. Ion–ion reactions can be used to assign charge states in very large systems [10]. It has been demonstrated that radical fragmentation can yield information about probable close contacts [11–17]. Various forms of spectroscopy can also be used to obtain information on biomolecules [18, 19]. Peptides and proteins are also amenable to examination by excitation energy-transfer (EET) where distance constraint information is obtained and used to guide simulations toward relevant structures [20–23].
The primary drawback for all of these gas-phase methods is that the target molecule is examined in isolation, without solvent or other interaction partners. Furthermore, the molecule must be removed from solvent and ionized (or allowed to retain a net charge) in order to be observed. Typically this is achieved with electrospray ionization (ESI); however, mechanistic details for ESI are not completely understood and it is unclear how protein structure evolves during electrospray, although recent progress has been made [24, 25]. Ultimately, results have been reported where native protein structure is either retained or lost upon examination in the gas phase [26, 27]. A bewildering number of experimental setups, conditions, and parameters can influence retention of native-like structure; however, one important difference between solution and the gas phase (which cannot be avoided regardless of experimental parameters) relates to solvation of charged functional groups. For example, in water, charged side chains of proteins such as lysine and arginine, or aspartic and glutamic acid, will interact strongly with water. If the solvent were somehow instantaneously removed (leaving the protein naked in the gas phase), the charged side chains would seek intramolecular solvation and potentially disrupt the structure [28].
Previous experiments have suggested that this structural reorganization can be prevented (or at least slowed down) if the solvent interaction is replaced with a surrogate. It has been well established that 18-crown-6 (18C6) interacts favorably with protonated amino acid side chains [29–31]. The interaction is strongest for lysine or the N-terminus, where three hydrogen bonds can form between each N–H and every other oxygen atom on the ring. The interaction is also strongest in the absence of readily available intramolecular hydrogen bonds or salt bridges [32]. Ion mobility results have demonstrated that partial solvation of the side chains by 18C6 allows higher charge states of ubiquitin to retain a more compact, native-like fold in the gas phase [33]. It was postulated that charge solvation by 18C6 prevented structural reorganization during transfer into the gas phase.
In the present work, we seek to examine in greater detail how 18C6 influences side-chain solvation and ultimately peptide structure. Action-EET spectra of several microsolvated and nonsolvated peptides are used in conjunction with molecular dynamics simulations in order to elucidate structural differences and the role 18C6 solvation plays in the transition from solution phase to gas phase.
Experimental
Materials
The peptide CEGNVRVSRELAGHTGY (GTP Gβ) was purchased from American Peptide Company (Sunnyvale, CA, USA). CNLYIQWLKDGGPSSGRPPPS (C-trpcage) was synthesized following standard solid-phase peptide synthesis procedure [34] and purified by HPLC. Propyl mercaptan (PM) used for disulfide formation was purchased from Acros Organics (Geel, Belgium).
Peptide Modification
All peptides were modified with PM prior to use via a procedure described previously [20]. Modification was followed by lyophilization to remove all solvent before resuspension in 10:90, 50:50, or 80:20 MeOH/H2O solutions. Prior to ESI, 2–4 uL of 1 mM 18C6 was added to the electrospray solution to a final volume of 500 μL and sprayed without further modification.
Photodissociation Mass spectrometry
Modified peptide solutions for action-EET experiments were sprayed through an ESI source into a Thermo Finnigan LTQ, which was modified to accept 266 nm Nd:YAG laser pulses (Continuum, Santa Clara, CA, USA) into the ion trap. UV-photodissociation experiments were performed by first isolating the ion of interest and exposing it to a single laser pulse before scanning out. The PD yield is defined as [product intensity/(product + precursor)] × 100. For most systems, the loss of PM is clean, yielding one significant product. If significant additional degradation of the cysteine side chain or complex occurs, this is included as well (for example, if UV irradiation resulted in a peak corresponding to loss of propyl mercaptan and crown, this loss would be included in the overall PD yield). An average of 50 scans was taken to generate the spectrum. A similar setup, used for collection of action spectra, consists of a Thermo LTQ XL that was modified to accept laser pulses from a Nd:YAG pumped optical parametric oscillator (OPO) laser (Opotek Inc., Carlsbad, CA, USA) set in the range of 250–300 nm. Again, after subsequent isolation of the parent ion, a single laser pulse excites ions before they are scanned out. This process is repeated over the entire range of 250–300 nm and yields for each of the collective spectra are calculated, also taking into account the laser power at each wavelength for normalization to the power of our 4 mJ 266 nm Nd:YAG. Once spectra are normalized to be representative of yields on a 266 nm 4 mJ Nd:YAG laser, they are finally plotted as a function of wavelength versus PD yield. A spline piecewise polynomial fit is used to construct solid lines in resulting action spectra.
Simulations
Molecular dynamics (MD) were performed using MacroModel [35] (Schrodinger Inc., Portland, OR, USA) with OPLS atomic force fields in order to estimate disulfide Tyr/Trp distances. Simulated annealing was carried out in two rounds; sampling at 1000 K was carried out to ensure adequate coverage of the structural space, followed by a second round of annealing at 450 K for selected structures to probe locally accessible conformations and better define the nearest minima. For runs at 1000 K, each of the 1000 obtained structures begins its cycle at 300 K with a 1.5 fs step time and 1 ps simulation time followed by a rise in temperature to 1000 K with a 1.0 fs step time and 10 ps simulation, then the temperature is gradually stepped back down to 750, 500, 300, and finally 200 K with step/simulation times of 1.5 fs and 10 ps, respectively. The step and simulation times for runs at 450 K are the same, but with the temperature ramping schedule being 300, 450, 300, and finally 200 K.
Results and Discussion
We have previously demonstrated that action-EET, where energy transfers from tyrosine or tryptophan to a disulfide bond, yields distance constraints useful for guiding simulations to determine peptide structure [20–22]. Briefly, a laser pulse causes electronic excitation of a chromophore, which then transfers energy to a cysteine residue capped with a disulfide bound propyl mercaptan (PM). The energy transfer leads to prompt dissociation of the disulfide (an easily detectable mass shift) and the intensity of the loss correlates with the distance between the chromophore and disulfide according to an exponential decay function. The derived distance constraint can be used to evaluate or guide molecular dynamics calculations to identify relevant structures.
GTP Peptide
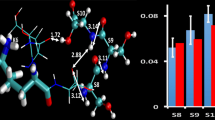
We examined GTP (CEGNVRVSRE LAGHTGY) with action-EET in the presence and absence of 18C6 and the results are shown in Figure 1. GTP contains an N-terminal cysteine and a C-terminal tyrosine. We have previously established that energy transfer from tyrosine to a disulfide is constrained to ~6 Å, meaning that fragmentation of the disulfide bond will only be enhanced for GTP if the three-dimensional structure brings the two termini into close proximity. The action spectrum for the 3+ charge state of GTP (Figure 1a) decays steadily from 250 to 300 nm. As reported previously, [21] this spectrum is consistent with minimal (though still non-zero) energy transfer from tyrosine. Interestingly, the addition of a single 18C6 leads to significant enhancement of energy transfer, suggesting the two ends of the peptide are brought closer when one of the charges is solvated. For the 2+ charge state (Figure 1b), the reverse trend is observed. The free peptide yields an action EET spectrum with distinctive tyrosine features (i.e., the double peaked absorption), and the degree of energy transfer is consistent with close proximity of the donor and acceptor. In contrast, addition of 18C6 to the 2+ charge state leads to a significant decrease in energy transfer, consistent with increasing the distance between the termini. The spectra in Figure 1 clearly demonstrates that solvation by 18C6 has a significant influence on the three-dimensional structures of both the 2+ and 3+ charge states.
We have also demonstrated previously that structural information can be obtained from the stability of the 18C6 adducts themselves [36]. Noncovalent interaction strength for 18C6 is a function of both the target residue (i.e., lysine interacts more strongly than arginine) and the potential for intramolecular solvation (i.e., for lysine on two different peptides, if one lysine can be easily intramolecularly solvated, it will form a less stable adduct). The PD spectrum for [GTP + 18C6 + 2H]2+ is shown in Figure 2a. In addition to cleavage of PM, significant 18C6 loss is detected. There is also a small amount of 18C6 and PM loss. In contrast, data for the 3+ charge state (Figure 2b) reveals minimal loss of 18C6 relative to PM. These results suggest the 18C6 adduct is more stable on the 3+ charge state and that the interaction site for 18C6 on the 2+ charge state is weaker by comparison. These results are further corroborated by other data from the mass spectra. The relative abundance in the full MS for the 2+ adduct relative to the 2+ peptide is 1:59, whereas the relative ratio is 1:5 for the 3+ ion, also consistent with greater stability for the 3+ adduct. We have also previously demonstrated that mass shifts relative to the expected mass for peptide/18C6 peaks result from adduct instability [36]. The mass shift observed for the 2+ adduct is 0.5 Da compared with 0.1 Da for the 3+ adduct, again consistent with reduced relative stability for the 2+ complex.
One thousand cycles of simulated annealing were used to investigate potential structures for these peptides. In order to more fully explore the structural space of these systems, annealing was first carried out at 1000 K to avoid kinetic trapping in local minima. For select structures, further simulated annealing was carried out at 450 K to allow for further structural refinement and evaluation of structural stability. This temperature was chosen to reflect mild heating that may occur during the electrospray process. For each charge state, all reasonable charge state isomers were examined. In other words, any charge state combination that yields the appropriate total charge via deprotonation of acidic sites and protonation of basic sites was investigated. It is important to note that comparison of the energetics between different classes of charge state isomers is not meaningful in MD calculations. For example the energies for isomers of the same peptide in protonated (+) versus doubly protonated/deprotonated (+-+) states is not possible because MD does not account for the possibility that the charge separated (zwitterionic) state may not be stable. Favorable Coulombic terms will always lead to lower energies for the charge separated conformers; however, these structures may not exist because of proton transfer (which is not allowed in MD calculations). Higher level theory capable of evaluating zwitterionic stability is not feasible for molecules of the size examined herein. In contrast, energies within a charge state class can be compared (e.g., single protonation at two different sites. In addition, for 18C6 adducts, calculations were carried out at all potential basic binding sites. In summary, a myriad of potential structures must be generated by theory, and guidance from experimental data is required to sort out potentially important structures.
Select results from simulated annealing for GTP 3+ are shown in Figure 3 (the complete set of results is found in the Supporting Information). The data are represented in scatter plots with the minimum distance between chromophores plotted on the x-axis and potential energy on the y-axis. The degree of structural similarity is represented in the color scale, which is a measure of the distance between the most sequence remote positively charged sites. After significant exploration, this indicator was found to be most representative of structural diversity, most likely due to the fact that charged sites considerably influence structure in the gas phase. The approximate region for experimentally consistent distances is shaded in blue.
The canonical way to obtain the 3+ charge state for GTP is by protonation of both arginine residues and the histidine (i.e., the R6R9H14 charge state isomer). Results for this isomer are shown in Figure 3a. The lowest energy structure has a Tyr/disulfide distance of ~9 Å. At this distance, no EET would be expected, and therefore it is clear that if the R6R9H14 charge state isomer is populated, some fraction of the experimentally observed ions have not rearranged to the lowest energy gas phase structure. Similar arguments apply to the other 3+ charge state isomers, indicating that regardless of charge state isomer, some fraction of GTP3+ is kinetically trapped in a structure not corresponding to the gas-phase minimum.
The experimental results for GTP3+ suggest weak EET. There are several potential scenarios that could explain this result. One possibility would be a structure or family of structures with donor/acceptor distances close to the EET cutoff distance. These structures would fall within the blue bands shown in Figure 3. Alternatively, a diverse population of structures where most have donor/acceptor distances greater than the EET cutoff but also including a small minority of the structures within the EET cutoff might produce the observed experimental results. Due to this ambiguity in the data, structures from all charge state isomers are potentially viable for GTP 3+, and it is difficult to further narrow down the results in this particular case. However, it can be concluded that the experimentally observed structures do not represent the lowest energy gas-phase conformations. Furthermore, it is still clear from the experimental results that solvation by 18C6 influences the structure significantly.
Selected results from simulations on GTP3+ with 18C6 are shown in Figure 4, and the remaining results are found in the Supporting Information. 18C6 stability (as detailed above) suggests strong binding, making the N-terminus the most likely binding site. The (nterm)R6R9 and (nterm)R6R9E10H14 charge state isomers, where parentheses are used to denote 18C6 binding sites, yield lowest energy isomers outside the experimentally consistent region (see Figure 4a and c). In contrast, the lowest energy structures for (nterm)E2R6R9H14 are within the experimentally determined energy transfer window (Figure 4b). The distribution for another conformer, (nterm)R6R9E10H14, is significantly biased away from the experimentally consistent region, and simulations at 450 K indicate that any kinetic barriers that might retain structures in the experimentally consistent region are small. This charge state isomer is, therefore, less likely to be populated.
Computational analysis of GTP2+ is stymied relative to GTP3+ by the existence of a greater number of potential charge state isomers. The experimental data is most clear for the naked 2+ charge state, which exhibits a high level of EET and a distinct shoulder at ~283 nm, consistent with no hydrogen bonding or charge proximity to the tyrosine side chain. These results are most consistent with simulations for the R6R9E10H14 isomer, which has a structural family with the appropriate donor/acceptor distance that remains kinetically trapped at 450 K. Unfortunately, this ion does not reveal information about solvation by 18C6, which is the focus of this manuscript. It is clear the 18C6 solvated 2+ ion is significantly different in structure; however, many possibilities from the simulations are consistent with experiment. These results can be found in the Supporting Information.
C-Trpcage
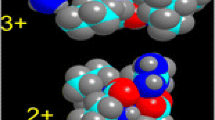
Trpcage is an engineered mini-protein with a well-known structure [37]. (CNLYIQWLKD GGPSSGRPPPS, the N-terminal cysteine is added in our experiment to enable action-EET). Trpcage was designed to fold quickly in water, yet previous work has shown that the 2+ charge state retains features consistent with the native fold in the gas phase [19, 20]. However previous work also yielded conflicting results regarding the presence and location of salt bridges in the 2+ structure [38, 39]. Our previous results for the 3+ charge state revealed significantly reduced EET relative to the 2+ charge state. A salt bridge-supported structure is not very feasible in the 3+ charge state (all acidic sites become neutral, leaving no room for counter charges); therefore, a salt bridge was suggested to account for the difference. Interestingly, EET increases dramatically when 18C6 is attached to the 3+ charge state, as shown in Figure 5a. This spectrum is similar to that of 2+ unsolvated C-Trpcage (Figure 5d), and contains contributions of EET from both Tyr and Trp.
(a) Action-EET spectrum of C-Trpcage 3+(purple) versus C-Trpcage 3+ + crown (red). (b) 450 K annealing starting with native structure for 3+ C-Trpcage solvated by crown on N-terminus. (c) 1000 K sampling of C-Trpcage 3+ crown starting from the native structure. (d) Action-EET spectrum of C-Trpcage 3+ crown (red) compared to the C-Trpcage 2+ action spectrum (purple)
Attempts were also made to examine 18C6 solvation of the 2+ charge state; however, it was not possible to generate a stable 18C6 adduct for the 2+ charge state. This is consistent with previous results where it was shown that the presence of salt bridges interferes with the formation of 18C6 adducts [32]. However, this interference would be expected if either the N-terminus or lysine residue was involved in salt bridges.
Simulated annealing of C-Trpcage was carried out at 450 K starting from the known solution-phase structure in order to mimic the process occurring during gentle electrospray. Interestingly, the naked 3+ ion is kinetically trapped under these conditions with fairly short Tyr/disulfide distances (the vast majority are <6 Å, see Supporting Information S6). At first glance, these results appear to be inconsistent with the experimental data; however, closer inspection of the relevant structures reveals that the protonated N-terminus is placed directly in between the tyrosine residue and the PM-modified cysteine (see Figure 6a). Placing a charged group directly between donor and acceptor would be expected to significantly interfere with EET. Therefore, even though the Tyr/disulfide distances from calculations are short, they can be reasonably considered to be consistent with the experimental data. These results suggest that the location of charged groups should be taken into consideration for analysis of EET experimental data.
(a) Unsolvated C-trpcage 3+ structure. (b) C-trpcage 3+ crown structure solvated at N-terminus. Note inverted orientation of protonated N-terminus relative to tyrosine and tryptophan. Lysine, aspartic acid, and arginine residues are highlighted in pink whereas tryptophan and tyrosine are denoted in orange
Simulated annealing on solvated C-Trpcage was explored by placing 18C6 on all three protonated sites (Supporting Information S6). Solvation of lysine and arginine yields structures where the protonated N-terminus remains located between the disulfide bond and the tyrosine residue, similar to the structure of the unsolvated system. In addition, the arginine solvated structures are significantly higher in energy, and the experimentally determined strength of 18C6 binding is not consistent with attachment at arginine. Therefore, binding at arginine is unlikely. Interestingly, a structural family with experimentally consistent donor/acceptor distances for both tyrosine and tryptophan was identified for 18C6 bound to the N-terminus (Figure 5b). These structures are within 20 kJ/mol of the lowest energy structures that were found by low temperature annealing and are lower in energy than any structure found by high temperature annealing (Figure 5c and Supporting Information S5). Examination of the structures within this family reveals that solvation by 18C6 replaces the intramolecular hydrogen bonding that drives the protonated N-terminus between the tyrosine and disulfide in the unsolvated structure (Figure 6b), which should restore energy transfer efficiency. Collisional activation of 3+ C-Trpcage-18C6 yields primarily unsolvated 3+ C-Trpcage, but also some of the unsolvated 2+ ion. Action-EET on the 3+ ion formed by CID yields nearly identical results to those for 3+ C-Trpcage sampled directly (Supporting Information S7). These results are consistent with the proposed structures because simple rotation of the N-terminus after loss of 18C6 would yield the unsolvated structure. Furthermore, loss of 18C6 from lysine or arginine would not be expected to result in a significant energy transfer change, further supporting the conclusion that these sites are not where 18C6 attaches to C-Trpcage. Another interesting result from these CID experiments is that action-EET of the 2+ ion formed by 18C6 loss from C-Trpcage 3+ also shows nearly identical energy transfer efficiency to the unsolvated 2+ ion when sampled directly (Supporting Information S8). Given that abstraction of a proton from the N-terminus (or even Lys) would not be expected to restore the Lys-Asp-Arg salt bridge for the newly formed 2+ ion, it is possible that retention of the salt bridge is not necessary for Trpcage stability as previously reported.
Conclusion
These experiments demonstrate that 18C6 microsolvation does influence gas-phase structure. Many of the systems examined herein are clearly not consistent with global gas-phase minima, suggesting that significant kinetic trapping occurs during the electrospray process. Solvation of available charges by 18C6 reduces the need for intramolecular solvation (and the structural changes that accompany it) upon solvent removal. Given that these results were obtained on small systems, which are most prone to structural rearrangements [40], it is likely that microsolvation will similarly stabilize native-like structures in larger proteins, as suggested previously [33].
References
Konijnenberg, A., Butterer, A., Sobott, F.: Native ion mobility-mass spectrometry and related methods in structural biology. Biochim. Biophys. Acta 1834, 1239–1256 (2013)
Lössl, P., Snijder, J., Heck, A.J.R.: Boundaries of mass resolution in native mass spectrometry. J. Am. Soc. Mass Spectrom. 25, 906–917 (2014)
Bleiholder, C., Dupuis, N.F., Wyttenbach, T., Bowers, M.T.: Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to β-sheet in amyloid fibril formation. Nat. Chem. 3, 172–177 (2011)
Bohrer, B.C., Merenbloom, S.I., Koeniger, S.L., Hilderbrand, A.E., Clemmer, D.E.: Biomolecule analysis by ion mobility spectrometry. Annu. Rev. Anal. Chem. (Palo Alto, CA) 1, 293–327 (2008)
Do, T.D., LaPointe, N.E., Nelson, R., Krotee, P., Hayden, E.Y., Ulrich, B., Quan, S., Feinstein, S.C., Teplow, D.B., Eisenberg, D., Shea, J.-E., Bowers, M.T.: Amyloid β-protein C-terminal fragments: formation of cylindrins and β-barrels. J. Am. Chem. Soc. 138, 549–557 (2016)
Allison, T.M., Reading, E., Liko, I., Baldwin, A.J., Laganowsky, A., Robinson, C.V.: Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat. Commun. 6, 8551 (2015)
Shepherd, D.A., Marty, M.T., Giles, K., Baldwin, A.J., Benesch, J.L.P.: Combining tandem mass spectrometry with ion mobility separation to determine the architecture of polydisperse proteins. Int. J. Mass Spectrom. 377, 663–671 (2015)
Marklund, E.G., Degiacomi, M.T., Robinson, C.V., Baldwin, A.J., Benesch, J.L.P.: Collision cross sections for structural proteomics. Structure 23, 791–799 (2015)
Tian, Y., Han, L., Buckner, A.C., Ruotolo, B.T.: Collision induced unfolding of intact antibodies: rapid characterization of disulfide bonding patterns, glycosylation, and structures. Anal. Chem. 87, 11509–11515 (2015)
Laszlo, K.J., Bush, M.F.: Analysis of native-like proteins and protein complexes using cation to anion proton transfer reactions (CAPTR). J. Am. Soc. Mass Spectrom. 26, 2152–2161 (2015)
Ly, T., Julian, R.R.: Residue-specific radical-directed dissociation of whole proteins in the gas phase. J. Am. Chem. Soc. 130, 351–358 (2008)
Sun, Q., Nelson, H., Ly, T., Stoltz, B.M., Julian, R.R.: Side chain chemistry mediates backbone fragmentation in hydrogen deficient peptide radicals. J. Proteome Res. 8, 958–966 (2009)
Diedrich, J.K., Julian, R.R.: Site-specific radical directed dissociation of peptides at phosphorylated residues. J. Am. Chem. Soc. 130, 12212–12213 (2008)
Ly, T., Julian, R.R.: Tracking radical migration in large hydrogen deficient peptides with covalent labels: facile movement does not equal indiscriminate fragmentation. J. Am. Soc. Mass Spectrom. 20, 1148–1158 (2009)
Moore, B.N., Ly, T., Julian, R.R.: Radical conversion and migration in electron capture dissociation. J. Am. Chem. Soc. 133, 6997–7006 (2011)
Moore, B.N., Julian, R.R.: Dissociation energies of X–H bonds in amino acids. Phys. Chem., Chem. Phys. 14, 3148 (2012)
Tureček, F., Julian, R.R.: Peptide radicals and cation radicals in the gas phase. Chem. Rev. 113, 6691–6733 (2013)
Talbot, F.O., Rullo, A., Yao, H., Jockusch, R.A.: Fluorescence resonance energy transfer in gaseous, mass-selected polyproline peptides. J. Am. Chem. Soc. 132, 16156–16164 (2010)
Iavarone, A.T., Parks, J.H.: Conformational change in unsolvated Trp-cage protein probed by fluorescence. J. Am. Chem. Soc. 127, 8606–8607 (2005)
Hendricks, N.G., Lareau, N.M., Stow, S.M., Mclean, J.A., Julian, R.R.: Bond-specific dissociation following excitation energy transfer for distance constraint determination in the gas phase. J. Am. Chem. Soc. 136, 13363–13370 (2015)
Hendricks, N.G., Julian, R.R.: Characterizing gaseous peptide structure with action-EET and simulated annealing. Phys. Chem., Chem. Phys. 17, 25822–25827 (2015)
Hendricks, N.G., Julian, R.R.: Two-step energy transfer enables use of phenylalanine in action-EET for distance constraint determination in gaseous biomolecules. Chem. Commun. 51, 12720–12723 (2015)
Daly, S., Poussigue, F., Simon, A., MacAleese, L., Bertorelle, F., Chirot, F., Antoine, R., Dugourd, P.: Action-FRET: Probing the molecular conformation of mass-selected gas-phase peptides with förster resonance energy transfer detected by acceptor-specific fragmentation. Anal. Chem. 86, 8798–8804 (2014)
McAllister, R.G., Metwally, H., Sun, Y., Konermann, L.: Release of native-like gaseous proteins from electrospray droplets via the charged residue mechanism: insights from molecular dynamics simulations. J. Am. Chem. Soc. 137, 12667–12676 (2015)
Yue, X., Vahidi, S., Konermann, L.: Insights into the mechanism of protein electrospray ionization from salt adduction measurements. J. Am. Soc. Mass Spectrom. 25, 1322–1331 (2014)
Skinner, O.S., McLafferty, F.W., Breuker, K.: How ubiquitin unfolds after transfer into the gas phase. J. Am. Soc. Mass Spectrom. 23, 1011–1014 (2012)
Wyttenbach, T., Bowers, M.T.: Structural stability from solution to the gas phase: native solution structure of ubiquitin survives analysis in a solvent-free ion mobility-mass spectrometry environment. J. Phys. Chem. B 115, 12266–12275 (2011)
Breuker, K., McLafferty, F.W.: Stepwise evolution of protein native structure with electrospray into the gas phase, 10(–12) to 10(2) s. Proc. Natl. Acad. Sci. U.S.A. 105, 18145–18152 (2008)
Julian, R.R., Beauchamp, J.: Site specific sequestering and stabilization of charge in peptides by supramolecular adduct formation with 18-crown-6 ether by way of electrospray ionization. Int. J. Mass Spectrom. 210/211, 613–623 (2001)
Ly, T., Julian, R.R.: Using ESI-MS to probe protein structure by site-specific noncovalent attachment of 18-crown-6. J. Am. Soc. Mass Spectrom. 17, 1209–1215 (2006)
Chen, Y., Rodgers, M.T.: Structural and energetic effects in the molecular recognition of protonated peptidomimetic bases by 18-crown-6. J. Am. Chem. Soc. 134, 2313–2324 (2012)
Liu, Z., Cheng, S., Gallie, D.R., Julian, R.R.: Exploring the mechanism of selective noncovalent adduct protein probing mass spectrometry utilizing site-directed mutagenesis to examine ubiquitin. Anal. Chem. 80, 3846–3852 (2008)
Warnke, S., Von Helden, G., Pagel, K.: Protein structure in the gas phase: the influence of side-chain microsolvation. J. Am. Chem. Soc. 135, 1177–1180 (2013)
Chan, W.C., White, P.D.: Fmoc Solid Phase Peptide Synthesis, 1st edn. Oxford University Press, New York (2000)
Maestro, ver. 9.7. Schrödinger, LLC: New York, NY (2012)
Tao, Y., Julian, R.R.: Factors that influence competitive intermolecular solvation of protonated groups in peptides and proteins in the gas phase. J. Am. Soc. Mass Spectrom. 24, 1634–1640 (2013)
Neidigh, J.W., Fesinmeyer, R.M., Andersen, N.H.: Designing a 20-residue protein. Nat. Struct. Biol. 9, 425–430 (2002)
Kjeldsen, F., Silivra, O.A., Zubarev, R.A.: Zwitterionic states in gas-phase polypeptide ions revealed by 157-nm ultra-violet photodissociation. Chem. Eur. J. 12, 7920–7928 (2006)
Adams, C.M., Kjeldsen, F., Zubarev, R.A., Budnik, B.A., Haselmann, K.F.: Electron capture dissociation distinguishes a single D-amino acid in a protein and probes the tertiary structure. J. Am. Soc. Mass Spectrom. 15, 1087–1098 (2004)
Hamdy, O.M., Julian, R.R.: Reflections on charge state distributions, protein structure, and the mystical mechanism of electrospray ionization. J. Am. Soc. Mass Spectrom. 23, 1–6 (2012)
Acknowledgments
The authors thank the NSF for financial support (CHE-1401737).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 4813 kb)
Rights and permissions
About this article
Cite this article
Bonner, J.G., Hendricks, N.G. & Julian, R.R. Structural Effects of Solvation by 18-Crown-6 on Gaseous Peptides and TrpCage after Electrospray Ionization. J. Am. Soc. Mass Spectrom. 27, 1661–1669 (2016). https://doi.org/10.1007/s13361-016-1456-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-016-1456-3