Abstract
Xenografts are commonly used to test the effect of new drugs on human cancer. However, because of their heterogeneity, analysis of the results is often controversial. Part of the problem originates in the existence of tumor cells at different metabolic stages: from metastatic to necrotic cells, as it happens in real tumors. Imaging mass spectrometry is an excellent solution for the analysis of the results as it yields detailed information not only on the composition of the tissue but also on the distribution of the biomolecules within the tissue. Here, we use imaging mass spectrometry to determine the distribution of phosphatidylcholine (PC), phosphatidylethanolamine (PE), and their plasmanyl- and plasmenylether derivatives (PC-P/O and PE-P/O) in xenografts of five different tumor cell lines: A-549, NCI-H1975, BX-PC3, HT29, and U-87 MG. The results demonstrate that the necrotic areas showed a higher abundance of Na+ adducts and of PC-P/O species, whereas a large abundance of PE-P/O species was found in all the xenografts. Thus, the PC/PC-ether and Na+/K+ ratios may highlight the necrotic areas while an increase on the number of PE-ether species may be pointing to the existence of viable tumor tissues. Furthermore, the existence of important changes in the concentration of Na+ and K+ adducts between different tissues has to be taken into account while interpreting the imaging mass spectrometry results.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Xenografts are a useful model to evaluate the pathobiology of tumors and to investigate the response to new antitumor drug candidates for human treatment [1, 2]. They are commonly developed in immunocompromised (nude) mice by injecting human tumor cells subcutaneously or onto a specific tissue in which the tumor originated [3]. One of the main advantages of this model is the possibility of evaluating the response to the treatment in human-originated cells and in a more physiological context than a culture dish. However, within a same experiment, xenografts usually differ in their morphology, growth rate, and vascularization level. For example, a rapid growth may lead to a poor vascularization of the tissue, which in turn may result in a generalized hypoxic state and, eventually, in cell death. That is, as it happens in clinical tumors, xenografts are formed by cells at different physiological stages, exposed to slightly different environments depending on their distance to the blood vessels, hindering altogether the comparison between samples [4–6].
An attractive approach to identify a valid biomarker that would correlate with the physiological stage of tumor cells may be to analyze the distribution of metabolites within xenograft samples by imaging mass spectrometry (IMS). Some attempts to find such biomarkers by using IMS were carried out, but the results were limited to a single type of xenograft and may not hold for xenografts from different cell lines [7, 8]. On the basis of those preliminary works, we explored here the predictive ability of the most abundant ether glycerophospholipids (GP) (i.e., plasmanyl phosphatidylcholine (PC-O), plasmenyl PC (PC-P), plasmanyl phosphatidylethanolamine (PE-O), and plasmenyl PE (PE-P) lipid species, Scheme 1), as biomarkers related to the physiological stages of tumor cells constituting the xenografts.
Ether GP species differ from the more common diacyl subclass in having a fatty alcohol rather than a fatty acid at the sn-1 position. The fatty alcohols are linked by a 1-O-alkyl ether bond, also termed a plasmanyl GP, or by a 1-O-(1Z-alkenyl) bond, containing a vinyl ether, termed a plasmenyl GP or plasmalogen (Scheme 1). In general, 1-O-alkyl groups are more abundant in choline GP, whereas 1-O-(1Z-alkenyl) groups are found primarily in ethanolamine GP. Ether GP synthesis is rather complex, starting in the peroxisomes and finishing in the endoplasmic reticulum [9]. Their physiological roles have been challenging to identify and are likely to be particular to different tissues, metabolic processes, and developmental stages [9]. Because of the absence of a carbonyl group at the sn-1 position, ether GPs have a higher propensity to form an inverse hexagonal phase (non-bilayer forming), which is a requirement for membrane fusion. Interestingly, the sn-2 position is frequently esterified with polyunsaturated fatty acids, particularly arachidonic and docosahexaenoic acids. Therefore, ether GP may be seen as reservoirs for these biologically active lipid mediators, released upon PLA2 hydrolysis. However, the most frequent role associated to ether GP is as an antioxidant, as it is generally accepted that the hydrogen atoms adjacent to the vinyl ether bond are preferentially oxidized over diacyl GP when exposed to various free radicals and singlet oxygen [10].
In a quest for a more universal set of biomarkers, here we describe the use of IMS to map the distribution of PC, PE, and their ether derivatives in xenografts of five different tumor cell lines: A-549 (adenocarcinomic human alveolar basal epithelial cells), BX-PC3 (human pancreatic cancer), HT29 (human colon carcinoma), NCI-H1975 (human lung cancer), and U-87 MG (human glioblastoma-astrocytoma). In agreement with previous works, we report an increased presence of ether PC and ether PE in all xenografts compared with non-tumor tissue [11–13]. As expected, they all showed very different lipid composition and relative abundance of species, owing to their different origin. However, as it will be demonstrated below, some common patterns in the distribution of the ether species were found across all the xenografts analyzed, revealing that their abundance and distribution may be a good indicator not only of the presence of neoplastic lesions but also of the existence of necrotic areas.
Materials and Methods
Reagents
Water, methanol, acetonitrile, 2-propanol and formic acid (Fisher Scientific, Fair Lawn, NJ, USA) were of Optima LC/MS grade. Leucine enkephalin acetate hydrate, ammonium acetate, and sodium hydroxide solution were purchased from Sigma-Aldrich Chemie (Steinheim, Germany).
Lipid standards LPC, (lysophosphatidylcholine synthetic), PE (liver, bovine), and PC (liver, bovine) were purchased from Avanti Polar-Lipids, Inc. (Alabaster, Alabama, USA). All stock standard solutions (in concentration 1000 μg/mL) were prepared in chloroform, and all were stored at −20°C in darkness. For identification purposes, reference solutions of each lipid class in a concentration 100 μg/mL were prepared in methanol-chloroform (2:1, v/v).
Chloroform and methanol used for xenograft lipid extraction were HPLC grade ACS ISO UV–VIS purity (Scharlau, Barcelona, Spain).
Animals and Tumor Grafts
Male NUDE (Swiss) Crl:NU (Ico)-Foxn1nu mice (5 wk-old, 30–35 g, Charles River Laboratories, Paris, France) were maintained in a thermostat cabinet (28°C; EHRET, Labor-U-Pharmatechnik) with a sterile air flow at a relative humidity of 40%–60% and a 12 h dark/light cycle. The human cancer cell lines (A-549, BX-PC3, HT-29, NCI-H1975, and U-87 MG) were obtained from the European Collection of Human Cell Cultures (ECACC, Salisbury, UK). Cells were cultured at 37°C in 5% CO2 and RPMI-1640 cell culture media supplemented with 10% fetal bovine serum (v∕v), 1 mM sodium pyruvate, 100 units∕mL penicillin, 0.1 mg∕mL streptomycin, and 0.25 μg∕mL amphotericin B. Media and other culture reagents were obtained from Sigma-Aldrich (Madrid, Spain). To obtain tumor xenografts, 7.5 × 106 cells (for NCI-H1975 and A-549 cell derived xenografts) or 107 cells (BX-PC3, HT-29, and U-87 MG derived xenografts) were inoculated subcutaneously into the animal dorsal area and after 1 wk, tumors were already visible. After the final tumor volume measurements, mice were sacrificed by decapitation. Then, the tumors were removed and immediately frozen in liquid nitrogen before being stored at −80°C. The tissue sections were not washed or manipulated in any way to preserve the original adduct concentration, which was very relevant to the study (see below). All experiments were carried out in accord with the animal welfare guidelines of the European Union and the Institutional Committee for Animal Research of the University of the Balearic Islands. Sections of ~15 μm thicknesses were prepared using no embedding material in cryostat (Leica CM3050S) at –20°C and placed on plain glass microscope slides. Sections were stored at –80°C until analysis.
Tissue Staining with Acridine Orange
As a control of tissue integrity, xenograft sections were first incubated with 4′,6-diamidino-2-phenylindole (DAPI; Dako, Barcelona, Spain) for 1 min at room temperature to stain cell nuclei. After washing three times with 0.1 M phosphate buffer solution (PBS, NaH2PO4 20 mM, Na2HPO4 80 mM, pH 7.4), sections were incubated 1 min at RT with a freshly prepared acridine orange solution (Sigma, Madrid, Spain) at 0.6 μg/mL in PBS. Finally, sections were washed three times with PBS, and a small drop of fluorescent mounting medium (Dako) was added onto the section. Images were obtained with Axioscope Cell Observer microscope (Carl Zeiss, Goettingen, Germany).
Sample Preparation for Imaging
During this work, between three and 20 sections from each xenograft were scanned, both in positive- and negative-ion modes, at spatial resolutions between 100 and 200 μm, using the Orbitrap analyzer of an LTQ-Orbitrap XL (ThermoFisher, San Jose, CA, USA), equipped with an N2 laser (100 μJ max power, elliptical spot of ~40 × 160 μm, 60 Hz repetition rate) [14]. Mass resolution was set to 100,000 at m/z = 400 Da to record the data, and the scanning range was 400–1000, both in positive- and negative-ion modes. MBT (2-mercaptobenzothiazole) [15] was used as matrix for positive- and negative-ion mode, and was deposited with the aid of a glass sublimator (Ace Glass 8023; Vineland, NJ, USA) [13]. The amount of matrix deposited over the section was a compromise between obtaining a uniform, thin enough layer so the matrix peaks did not interfere with the detection of lipids and depositing a sufficient amount of matrix to allow scanning for several hours [14].
The spectra were analyzed using dedicated software (MSIAnalyst; NorayBioinformatics S. L., Zamudio, Spain). During parsing, the size of the data was reduced, eliminating all the peaks whose intensity was lower than the 0.5% of the most intense peak on the spectrum, and the spectra were normalized using a total ion current algorithm [16]. Spectra were also aligned using the Xiong et al. method [17] and assuming a maximum misalignment of 0.02 Da., which is very conservative for an Orbitrap analyzer. During graphical representation, no interpolation or smoothing algorithms or any de-noising procedure was used, always trying to maintain the original aspect of the data.
Statistical analysis to identify the different areas in the sections was carried out using principal component analysis (PCA) [18] and k-means [19]. However, only the latter analysis will be presented here, as the clustering algorithm produced superior results.
Lipid identification in MALDI-IMS was based on a direct comparison between the value of the m/z and the lipids in the software’s lipid data base (>33,000 species plus their adducts) and those in the lipid maps data base (www.lipidmaps.org). Mass accuracy was always better than 10 ppm and it was typically better than 3 ppm. In those cases where no unequivocal assignment was found, a comparison with the data from UHPLC was performed. If any of the candidates were not detected by UHPLC-MS, and it was not detected as other adduct in the MALDI-IMS experiment, such candidate was removed from the list. Data from LC-MS/MS were also used to provide the identity of the acyl/alkyl substituents of the species detected (see Tables 1 and 2)
Lipid Extracts for UHPLC-MS
Lipids were extracted by the Bligh and Dyer method [20], applied as described elsewhere [13]. Briefly, approximately 90 mg of frozen tissue was homogenized with a tissue blender (Ultra-Turrax; Janke and Kunkel, Saufen, Germany). In each sample, 3.75 mL of 1:2 (v/v) CHCl3:MeOH was added and vortexed thoroughly for 1 min. Then, 1.25 mL of CHCl3 was added and the solution was also vortexed thoroughly for 1 min. Finally, 1.25 mL of distilled H2O was added and again the solution was vortexed thoroughly for 30 s. Samples were centrifuged at 1000 g for 5 min at 4°C to obtain a two-phase system (aqueous top, organic bottom) and the bottom phase was recovered. The latter was re-extracted by adding 2 mL of 1:2 (v/v) CHCl3:MeOH 1:1 (v/v) and 2 mL of H2O, and again vortexed thoroughly for 1 min. Samples were centrifuged at 1000 g for 10 min at 4°C. The lower phase was combined with the first organic phase, evaporated under N2 stream. Lipid extract was resuspended in n-hexane:isopropanol (3:2, v/v) and stored at −80°C until analysis.
UHPLC-MSE Analysis
The procedure used in this work was similar to that in reference [13]. Ultrahigh performance liquid chromatography (UHPLC) was carried out by using an ACQUITY UPLC system from Waters (Milford, MA, USA). Lipid extracts from the xenografts were injected onto a column (Acquity UPLC HSS T3 1.8 μm, 100 × 2.1 mm; Waters), which was heated to 65°C. Mobile phases consisted of acetonitrile and water with 10 mM ammonium acetate (40:60, v/v, phase A) and acetonitrile and isopropanol with 10 mM ammonium acetate (10:90, v/v, phase B). Separation was carried out in 13 min under the following conditions: 0–10 min, linear gradient from 40% to 100% B; 10–11 min, 100% B; and finally, re-equilibration of the system with 40% B (v/v) for 2 min prior to the next injection. Flow rate was 0.5 mL/min and injection volume was 5 μL. All samples were kept at 4°C during the analysis.
All UHPLC-MSE data were acquired on a SYNAPT G2 HDMS, with a quadrupole time of flight (Q-ToF) configuration, (Waters) equipped with an electrospray ionization (ESI) source that can be operated in both positive- and negative-ion modes. The capillary voltage was set to 0.7 kV (ESI+) or 0.5 kV (ESI−). Nitrogen was used as desolvation and cone gas, at flow rates of 900 L/h and 30 L/h, respectively. The source temperature was 120°C, and the desolvation temperature was 400°C.
Before analysis, the mass spectrometer was calibrated with a sodium formate 0.5 mM solution. Leucine-enkephalin solution (2 ng/μL) in acetonitrile:water (50:50, v/v + 0.1% formic acid) was utilized for the lock-mass correction of all the spectra and the ions at mass-to-charge ratios (m/z) 556.277 and 278.114, or 554.262 and 236.104, depending on the ionization mode, from this solution were monitored at scan time 0.3 s, and at 10 s intervals, three scans on average, using a mass window of ±0.5 Da. The rest of the conditions were: lockspary capillary 2.0 and 2.5 kV and collision energy 21 and 30 eV in ESI+ or ESI–, respectively. The reference internal calibrant was introduced into the lock mass sprayer at a constant flow rate of 10 μL/min using an external pump.
Data acquisition took place over the mass range 50–1200 Da in resolution mode (FWHM ≈ 20,000 at m/z = 956) with a scan time of 0.5 s and an inter-scan delay of 0.024 s. The mass spectrometer was operated in the continuum MSE acquisition mode for both polarities. During this acquisition method, the first quadrupole Q1 was operated in a wide band rf mode only, allowing all ions to enter the T-wave collision cell. Two discrete and independent interleaved acquisition functions were automatically created: the first function, typically set at 6 eV, collected low energy or data on unfragmented ions, whereas the second function collected high energy or data on fragmented ions, typically obtained by using a collision energy ramp from 15 to 40 eV. In both cases, Ar gas was used for collision induced dissociation (CID).
Data Processing for UHPLC-MSE and Identification of Lipids
Lipid analysis and identification followed a procedure similar to that in reference [13]. MassLynx software ver. 4.1 (Waters MS Technology, Mil-ford, MA, USA) was used for instrument control, data acquisition, and processing. For the MSE data processing, the software tool MSE Data Viewer (Waters MS Technologies, Manchester, UK) was utilized. The data generated by UHPLC-MSE were extracted using MSE Data Viewer (Waters MS Technologies). This software was used for aligning precursor and product-ion spectra according to retention times, generating an exportable text file, which was used for lipid identification using SimLipid software (Premier Biosoft, Palo Alto CA, USA).
The identification of lipids for which standards were available was carried out by the comparison of their retention times and MSE spectra recorded in positive- and negative-ion modes with those obtained by injecting standards in the same conditions. The identity of the rest of compounds was elucidated using the low collision energy MSE spectrum in positive- and negative-ion modes to determine the molecular weight. The higher collision energy MSE spectrum yielded the fragmentation and was used to elucidate other structural details. The software provided with the spectrometer allows one to correlate the fragmentation spectra with the corresponding parent ions. Some examples of chromatograms and fragmentation spectra may be found in the Supporting Information.
Results and Discussion
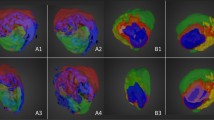
As demonstrated in previous works using IMS [1, 2, 13, 21–24], xenografts are highly heterogeneous samples, containing cells in different physiological stages. This is highlighted in the left column of Figure 1, where a representative section stained with acridine orange of each type of xenograft analyzed in the present work is shown. This type of staining has been widely used to assess DNA damage in many cell types [25]. When bound to double-stranded DNA, acridine orange has an emission maximum at 525 nm (green). However, when it associates with RNA, the maximum of the emission shifts to 650 nm (red). Consequently, the absence of green indicates that DNA integrity is compromised, and this can be associated to cell death. One must take into account that the colors are a relative scale and that the color scale may change between images. Therefore, whereas in some images necrotic areas appear as a bright red, like in HT-29 or NCI-H1975, in others like U-87 MG, they appear as a pale gold. The images demonstrate that the sections contained necrotic and viable tissue in different ratios. Several markers have been proposed to identify those areas, such as the presence of carnitine species or some phospholipids [7, 8]. These were tested in the present work, but none of them worked as a good marker of necrosis in all five xenograft types analyzed, although carnitine derivatives were the most promising ones.
Left column, acridine orange staining of representative sections of each type of xenograft studied in this work. Necrotic areas appear as yellow/orange or red. Center: [M + Na+]/([M + Na+] + [M + K+]) ratio for PC 32:0 and 32:1 along sequential sections extracted from an IMS experiment. Red color indicates a higher abundance of sodium adducts, whereas green indicates a higher abundance of potassium adducts. Right column, two (three in the case of BX-PC3) of the clusters found in the IMS experiment using k-means that overlap reasonably well with the viable and necrotic areas of the xenografts. Scale bars = 2 mm
Looking for a more universal marker of necrosis, we mapped the relative abundance of sodium and potassium in the tissue, as previous studies have reported changes in the abundance of those adducts during cell apoptosis and cancer [26]. To determine the Na+/K+ ratio it is necessary to find species whose adducts are free of other species interferences. Hence, we used here PC 32:0 and PC 32:1, as they are present in all the xenograft sections examined, and their Na+ and K+ adducts do not overlap with any known species identified by IMS or found in the extracts using LC/MS (m/z = 756.5519, 772.5259, 754.5363, and 770.5102 for PC 32:0 + Na+, PC 2:0 + K+, PC 32:1 + Na+, and PC 32:1 + K+, respectively). Thus, the central column in Figure 1 shows the [M + Na+]/([M + Na+] + [M + K+]) ratio found for PC 32:0 and PC 32:1 in consecutive sections to those stained with acridine orange, in a green-yellow-red color scale. In that scale, green indicates that potassium adducts are more abundant, whereas red indicates higher abundance of sodium adducts of the selected species. Comparison of the images generated in this way with the acridine orange staining show remarkable coincidences, taking into account that there are morphologic changes between sequential sections and that the staining is an aggressive procedure that may result in material loss. For example, the conjunctive tissue covering the xenograft [13] is usually lost during the staining.
Despite the morphologic changes, it is clear that those areas with necrotic tissue correlated well with a higher abundance of sodium adducts (red colors in the images of the central column). For example, the upper part of the HT-29 xenograft seemed to contain necrotic tissue, according to the acridine orange staining. Likewise, the concentration of Na+ adducts was higher in that part of the xenograft. Although less evident, the upper-left corner of U-87 MG and part of the central area present a golden color, indicative of the presence of non-viable tissue. In the Na+/K+ ratio image, those areas are also in red. Additionally, the tissue in the lower-left corner of the section appeared to have also higher sodium content, but that part of the tissue was lost during staining and, therefore, it did not show up.
Likewise, a good correlation between the abundance of Na+ adducts and the necrotic areas scattered along the A-549, those in the central part of NCI-H1975, and the necrotic lower part of BX-PC3 was found. Regarding the NCI-H1975, for some reason most of the tissue appears in red-orange color, which does not mean that it only contains necrotic areas but that the staining is more shifted towards the red than in other sections.
The right column in Figure 1 shows two of the clusters found in the IMS experiment using a k-means analysis, setting the number of clusters to four (five in the case of BX-PC3). Comparison with the other two columns show that the difference in the lipidome between necrotic and viable tissue was significant enough to result in two separated clusters, except for BX-PC3, in which the viable tissue was divided into two clusters, may be due to different populations of cells or to a different degree of vascularization of the tissue.
At this point, the question of the origin of the variation of the Na+/K+ ratio arises. Previous works demonstrated that the tissue damage produced during ischemia is accompanied by a deregulation of the Na+/K+-ATPase (or Na+ pump) inducing changes in the relative abundance of Na+/K+ adducts [27]. In the case of cancer, it is well known that tumor cells are usually in environments with lower pH than healthy tissue, probably owing to their unbalanced metabolism [28, 29]. The acid microenvironment will certainly have an effect on all ionic channels responsible for the maintenance of the strictly regulated intracellular milieu. In this context, it is known that different types of K+ channels are unequivocally associated with malignant progression [30, 31]. K+ channels can be modulated by a wide range of chemical and physical stimuli, including acidification [32–34] and hypoxia [35]. Interestingly, the pharmacologic inhibition of certain K+ channels exerted antitumor activity in certain cases [36], suggesting an increased channel activity in tumor cells compared with non-tumor cells [30, 31]. Such effect could explain the existence of a larger amount of potassium adducts in the viable areas of the xenograft, as PC species (usually in the outer leaflet of the plasma membrane) would find more potassium cations to interact with. Therefore, a higher amount of potassium may be a good marker for the viable areas of the tumor, whereas a higher abundance of sodium may be highlighting the necrotic/hypoxic areas. If this hypotheses is correct, it would also imply that the nature and concentration of adducts detected is linked to the original composition of the tissue, and this information is not lost during the desorption process. The results shown in Figure 1 have an additional and important consequence: the analysis of the changes in the lipid composition between different types of tissues determined by using IMS in positive-ion mode must take into account the adduct formation. Otherwise, significant errors may be made during data interpretation. This implies a previous identification of all the species representative of each tissue, to reduce their H+/Na+/K+ adducts to a single channel for each species, or to add salts (for example potassium salts) during matrix application to shift the cluster formation towards a single adduct.
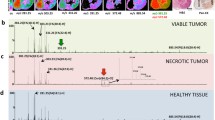
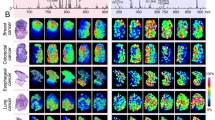
Regarding the distribution of PC/PE-ether species, Figure 2 shows the distribution of selected lipid species along the sections presented in Figure 1, whereas the average spectra may be found in Figure S1 of the Supporting Information. No PE-ether species were found in the IMS experiments over U87 MG or BX-PC3, probably due to interference with other PC species. It is clear from Figure 2 that the distribution of the selected species is not uniform along the sections selected and that the areas found during the clustering analysis reflect the xenograft heterogeneity.
Finally, we evaluated the correlation between the GP/GP-ether ratio and cell damage. A different PC/PC-ether ratio was also found in the areas of viable tissue compared with those with higher abundance of necrosis (Figure 3). Although a significant number of PC-ether species was found (Table 1), their concentration is always lower compared with the diacyl-PC species. One must take into account that structural species, such as PC 34:1, are usually the most intense peaks in the spectrum. Nevertheless, there was a significant and consistent increase in the concentration of PC-ether species in the necrotic areas compared with the viable tissue. The PE/PE-ether ratio was similarly studied. However, the number of PE species detected by IMS directly from the tissue was small, compared with the number of PC species. This can be due mostly to ion suppression effects but also because many PE species overlapped with the more abundant PC species. Consequently, their relative distributions could not be separated, unless MS/MS experiments were carried out directly on the section, which is only possible for components represented by peaks of relatively strong intensity, or running ion mobility experiments. Furthermore, PE species detected by IMS were divided between positive- and negative-ion modes and, therefore, it is hard to summarize all the species in the same figure, owing to normalization issues. Nevertheless, Figure S2 (Supplemental Material) shows the ratio found for a section of each type of xenograft. Whereas PE-ether species were found in higher abundance in HT29 necrotic areas, the opposite was found in necrotic areas of NCI-H1975, A-549, and U-87MG xenografts, indicating that either they are not good biomarkers for necrosis or that the number of species detected using MALDI-IMS was not representative enough to reach conclusions.
PC-ether/PC ratio in viable/necrotic tissue in each of the analyzed sections. The clusters in red were obtained using k-means (Figure 1). Two different regions of viable tissue were identified in the BX-PC3 sections
In fact, using LC/MS, a significantly larger number of PE-ether species was detected. Tables 1 and 2 collect the entire PC, PE, and their ether lipids species detected by this method, and Figure 4 represents the PE-ether/PE abundance ratio in extracts of each type of xenograft, determined using LC-MS/MS. There was a surprisingly high abundance of PE-ether species in every type of xenograft analyzed in this work, compared with normal tissue from the same organs, despite their different origin. Thus, whereas ether lipids are usually important components of brain or lung tissues [9], their abundance in colon (HT29) or pancreas (BX-PC3) was unexpected [9]. Consequently, if all the sections analyzed present such a high abundance of plasmalogens, it may be related to the tumor process. Importantly, previous publications demonstrated that the analysis of lipid extracts obtained from tumor human cells lines or human cancer biopsies showed increased levels of PC and/or PE ether lipids. In fact, these changes are correlated with malignancy and metastatic potential in various cancers [37–42]. Some studies demonstrated that plasmalogens are natural antioxidants, as the ether link is more prone to be oxidized than the double bonds usually found in many phospholipids in their fatty acid moiety [9, 43]. Thus, an increased abundance of ether species (PE ethers in this case) may be due to a protection mechanism used by the viable tumor cells and, therefore, the PE/PE - ethers ratio may be a good universal biomarker for the presence of malignant cells.
Conclusions
Here we have demonstrated the capabilities of IMS to map the lipid composition of five different types of xenografts generated from a wide variety of human tumor cell lines: lung (A-549 and NCI-H1975), brain (U-87 MG), pancreas (BX-PC3), and colon (HT29) by determining the distribution of PC, PE, and their ether derivatives. In good agreement with previous works, the results highlight the diverse microenvironments present in each xenograft, which very well resemble the heterogeneity of the clinical tumors, and that add a level of complexity to their study. Importantly, changes in the Na+/K+ relationship along the xenografts may be used to identify the necrotic areas, which showed higher abundance in sodium adducts. In addition, despite their heterogeneous origin, the analysis of the xenografts by HPLC-MS/MS showed a higher-than-expected abundance of both PC- and PE-ether species compared with normal tissue from the same organ. These molecules are well known to help cells to prevent double bonds oxidation, which is especially important under oxidative stress, as those usually found in tumors. Thus, our results point to the possibility of using the abundance of PE- and PC-ether species as biomarkers for the existence of neoplastic tissue. A further analysis by IMS analysis indicated that, in fact, PC-ether species were particularly enriched in necrotic areas compared with viable areas.
Finally, a word of caution when analyzing the results from IMS is required: the changes in the adduct concentration along the xenograft may lead to data misinterpretation if the effect is not taken into account during data analysis.
References
Fiebig, H.H., Maier, A., Burger, A.M.: Clonogenic assay with established human tumor xenografts: correlation of in vitro to in vivo activity as a basis for anticancer drug discovery. Eur. J. Cancer 40, 802–820 (2004)
Mischek, D., Steinborn, R., Petznek, H., Bichler, C., Zatloukal, K., Sturzl, M., Gunzburg, W.H., Hohenadl, C.: Molecularly characterized xenograft tumor mouse models: valuable tools for evaluation of new therapeutic strategies for secondary liver cancers. J. Biomed. Biotechnol. 2009, 437284 (2009)
Morton, C.L., Houghton, P.J.: Establishment of human tumor xenografts in immunodeficient mice. Nat. Protoc. 2, 247–250 (2007)
Huber, K., Feuchtinger, A., Borgmann, D.M., Li, Z., Aichler, M., Hauck, S.M., Zitzelsberger, H., Schwaiger, M., Keller, U., Walch, A.: A novel approach of MALDI drug imaging, immunohistochemistry, and digital image analysis for drug distribution studies in tissues. Anal. Chem. 86, 10568–10575 (2014)
Eirew, P., Steif, A., Khattra, J., Ha, G., Yap, D., Farahani, H., Gelmon, K., Chia, S., Mar, C., Wan, A., Laks, E., Biele, J., Shumansky, K., Rosner, J., McPherson, A., Nielsen, C., Roth, A.J.L., Lefebvre, C., Bashashati, A., de Souza, C., Siu, C., Aniba, R., Brimhall, J., Oloumi, A., Osako, T., Bruna, A., Sandoval, J.L., Algara, T., Greenwood, W., Leung, K., Cheng, H., Xue, H., Wang, Y., Lin, D., Mungall, A.J., Moore, R., Zhao, Y., Lorette, J., Nguyen, L., Huntsman, D., Eaves, C.J., Hansen, C., Marra, M.A., Caldas, C., Shah, S.P., Aparicio, S.: Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015)
Batubara, A., Carolan, V.A., Loadman, P.M., Sutton, C., Shnyder, S.D., Clench, M.R.: Thin-layer chromatography/matrix-assisted laser desorption/ionization mass spectrometry and matrix-assisted laser desorption/ionization mass spectrometry imaging for the analysis of phospholipids in ls174t colorectal adenocarcinoma xenografts treated with the vascular disrupting agent DMXAA. Rapid Commun. Mass Spectrom. 29, 1288–1296 (2015)
Chughtai, K., Heeren, R.M.A.: Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 110, 3237–3277 (2010)
Jiang, L., Chughtai, K., Purvine, S.O., Bhujwalla, Z.M., Raman, V., Pasa-Tolic, L., Heeren, R.M.A., Glunde, K.: MALDI-mass spectrometric imaging revealing hypoxia-driven lipids and proteins in a breast tumor model. Anal. Chem. 87, 5947–5956 (2015)
Braverman, N.E., Moser, A.B.: Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta – Mol. Basis of Dis. 1822, 1442–1452 (2012)
Broniec, A., Klosinski, R., Pawlak, A., Wrona-Krol, M., Thompson, D., Sarna, T.: Interactions of plasmalogens and their diacyl analogs with singlet oxygen in selected model systems. Free Radic. Biol. Med. 50, 892–898 (2011)
Fernandez, J.A., Ochoa, B., Fresnedo, O., Giralt, M.T., Rodriguez-Puertas, R.: Matrix-assisted laser desorption ionization imaging mass spectrometry in lipidomics. Anal. Bioanal. Chem. 401, 29–51 (2011)
Zemski Berry, K.A., Hankin, J.A., Barkley, R.M., Spraggins, J.M., Caprioli, R.M., Murphy, R.C.: MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 111, 6491–6512 (2011)
Fernández, R., Lage, S., Abad-García, B., Barceló-Coblijn, G., Terés, S., López, D.H., Guardiola-Serrano, F., Martín, M.L., Escribá, P.V., Fernández, J.A.: Analysis of the lipidome of xenografts using MALDI-IMS and UHPLC-ESI-QTOF. J. Am. Soc. Mass Spectrom. 25, 1237–1246 (2014)
Garate, J., Fernandez, R., Lage, S., Bestard-Escalas, J., Lopez, D.H., Reigada, R., Khorrami, S., Ginard, D., Reyes, J., Amengual, I., Barcelo-Coblijn, G., Fernandez, J.A.: Imaging mass spectrometry increased resolution using 2-mercaptobenzothiazole and 2,5-diaminonaphtalene matrices: application to lipid distribution in human colon. Anal. Bioanal. Chem. 407, 4697–4708 (2015)
Astigarraga, E., Barreda-Gomez, G., Lombardero, L., Fresnedo, O., Castano, F., Giralt, M.T., Ochoa, B., Rodriguez-Puertas, R., Fernandez, J.A.: Profiling and Imaging of lipids on brain and liver tissue by matrix-assisted laser desorption/ionization mass spectrometry using 2-mercaptobenzothiazole as a matrix. Anal. Chem. 80, 9105–9114 (2008)
Deininger, S.O., Ebert, M.P., Futterer, A., Gerhard, M., Rocken, C.: MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J. Proteome Res. 7, 5230–5236 (2008)
Xiong, X.C., Xu, W., Eberlin, L.S., Wiseman, J.M., Fang, X., Jiang, Y., Huang, Z.J., Zhang, Y.K., Cooks, R.G., Ouyang, Z.: Data processing for 3D mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 23, 1147–1156 (2012)
Wold, S., Esbensen, K., Geladi, P.: Principal component analysis. Chemometr. Intell. Lab. Syst. 2, 37–52 (1987)
Arthur, D., Vassilvitskii, S.: K-Means++: the advantages of careful seeding. In: Proceedings of the 18th Annual ACM-SIAM Symposium on Discrete Algorithms, New Orleans, 7–9 January 2007, pp. 1027–1035
Bligh, E.G., Dyer, W.J.: A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959)
Djidja, M.C., Francese, S., Loadman, P.M., Sutton, C.W., Scriven, P., Claude, E., Snel, M.F., Franck, J., Salzet, M., Clench, M.R.: Detergent addition to tryptic digests and ion mobility separation prior to MS/MS improves peptide yield and protein identification for in situ proteomic investigation of frozen and formalin-fixed paraffin-embedded adenocarcinoma tissue sections. Proteomics 9, 2750–2763 (2009)
Mattie, M., Christensen, A., Chang, M.S., Yeh, W., Said, S., Shostak, Y., Capo, L., Verlinsky, A., An, Z., Joseph, I., Zhang, Y., Kumar-Ganesan, S., Morrison, K., Stover, D., Challita-Eid, P.: Molecular characterization of patient-derived human pancreatic tumor xenograft models for preclinical and translational development of cancer therapeutics. Neoplasia 15, 1138–IN17 (2013)
Amstalden, V.H., Blackwell, T.R., Klinkert, I., Eijkel, G.B., Heeren, R.M.A., Glunde, K.: Multimodal mass spectrometric imaging of small molecules reveals distinct spatio-molecular signatures in differentially metastatic breast tumor models. Cancer Res. 70, 9012–9021 (2010)
Wu, Q., Li, M.y., Li, H.q., Deng, C.h., Li, L., Zhou, T.y., Lu, W.: Pharmacokinetic-pharmacodynamic modeling of the anticancer effect of erlotinib in a human non-small cell lung cancer xenograft mouse model. Acta Pharmacol. Sin. 34, 1427–1436 (2013)
Söderström, K., Parvinen, L., Parvinen, M.: Early detection of cell damage by supravital acridine orange staining. Experientia 33, 265–266 (1977)
Li, L., Li, P., Fang, J., Li, Q., Xiao, H., Zhou, H., Tang, B.: Simultaneous quantitation of Na+ and K+ in single normal and cancer cells using a new near-infrared fluorescent probe. Anal. Chem. 87, 6057–6063 (2015)
Hankin, J., Farias, S., Barkley, R., Heidenreich, K., Frey, L., Hamazaki, K., Kim, H.Y., Murphy, R.: MALDI mass spectrometric imaging of lipids in rat brain injury models. J. Am. Soc. Mass Spectrom. 22, 1014–1021 (2011)
Griffiths, J.R.: Are cancer cells acidic? Br. J. Cancer 64, 425–427 (1991)
Kato, Y., Ozawa, S., Miyamoto, C., Maehata, Y., Suzuki, A., Maeda, T., Baba, Y.: Acidic extracellular microenvironment and cancer. Cancer Cell Int. 13, 8 (2013)
Comes, N., Serrano-Albarrás, A., Capera, J., Serrano-Novillo, C., Condom, E., Ramón y Cajal, S., Ferreres, J.C., Felipe, A.: Involvement of potassium channels in the progression of cancer to a more malignant phenotype. Biochim. Biophys. Acta. 1848, 2477–2492 (2015)
Pardo, L.A., Stuhmer, W.: The roles of K+ channels in cancer. Nat. Rev. Cancer 14, 39–48 (2014)
Rajan, S., Wischmeyer, E., Xin Liu, G., Preisig-Müller, R., Daut, J., Karschin, A., Derst, C.: TASK-3, a novel tandem pore domain acid-sensitive K+channel: an extracellular histidine as pH sensor. J. Biol. Chem. 275, 16650–16657 (2000)
Duprat, F., Lesage, F., Fink, M., Reyes, R., Heurteaux, C., Lazdunski, M.: TASK, a human background K+ channel to sense external pH variations near physiological pH. EMBO J. 16, 5464–5471 (1997)
Kim, Y., Bang, H., Kim, D.: TASK-3, a new member of the tandem pore K+ channel family. J. Biol. Chem. 275, 9340–9347 (2000)
Buckler, K.J., Williams, B.A., Honore, E.: An oxygen-, acid- and anesthetic-sensitive TASK-like background potassium channel in rat arterial chemoreceptor cells. J. Physiol. Lond. 525, 135–142 (2000)
Gómez-Varela, D., Zwick-Wallasch, E., Knötgen, H., Sánchez, A., Hettmann, T., Ossipov, D., Weseloh, R., Contreras-Jurado, C., Rothe, M., Stühmer, W., Pardo, L.A.: Monoclonal antibody blockade of the human eag1 potassium channel function exerts antitumor activity. Cancer Res. 67, 7343–7349 (2007)
Fhaner, C.J., Liu, S., Ji, H., Simpson, R.J., Reid, G.E.: Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal. Chem. 84, 8917–8926 (2012)
Smith, R., Lespi, P., Di Luca, M., Bustos, C., Marra, F., Alaniz, M., Marra, C.: A reliable biomarker derived from plasmalogens to evaluate malignancy and metastatic capacity of human cancers. Lipids 43, 79–89 (2008)
Min, H., Lim, S., Chung, B., Moon, M.: Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal. Bioanal. Chem. 399, 823–830 (2011)
Dória, M.L., Cotrim, Z., Macedo, B., Simoes, C., Domingues, P., Helguero, L., Domingues, M.R.: Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res. Treat. 133, 635–648 (2012)
Merchant, T.E., Kasimos, J.N., de Graaf, P.W., Minsky, B.D., Gierke, L.W., Glonek, T.: Phospholipid profiles of human colon cancer using 31P magnetic resonance spectroscopy. Int. J. Colorectal Dis. 6, 121–126 (1991)
Dueck, D., Chan, M., Tran, K., Wong, J., Jay, F., Littman, C., Stimpson, R., Choy, P.: The modulation of choline phosphoglyceride metabolism in human colon cancer. Mol. Cell. Biochem. 162, 97–103 (1996)
Zoeller, R.A., Lake, A.C., Nagan, N., Gaposchkin, D.P., Legner, M.A., Lieberthal, W.: Plasmalogens as endogenous antioxidants: somatic cell mutants reveal the importance of the vinyl ether. Biochem. J. 338, 769–776 (1999)
Acknowledgments
This study was partially supported by the Institute of Health Carlos III (IHCIII, Ministerio de Economía y Competitividad), and the EC (European Regional Development Fund, ERDF, CP12/03338), by UPV/EHU (UFI 11/23) and by the Basque Government (consolidated groups). Technical support and personnel provided by the Servicio de Lipidómica of the SGIKER (UPV/EHU, MICINN, GV/E.G., ESF) is gratefully acknowledged. J.G. thanks the Basque Government for a pre-doctoral fellowship. G.B.-C. holds a ‘Miguel Servet’ contract from the IHCIII, Ministerio de Economía y Competitividad. S.T. holds a contract from the INSERM (Institut National de la santé de la recherche médical) Délégation regional de Bordeaux. F.G-S holds a contract from the Asociación Española Contra el Cáncer (AECC).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1951 kb)
Rights and permissions
About this article
Cite this article
Fernández, R., Garate, J., Lage, S. et al. Identification of Biomarkers of Necrosis in Xenografts Using Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom. 27, 244–254 (2016). https://doi.org/10.1007/s13361-015-1268-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1268-x