Abstract
Disulfide bonds are a post-translational modification (PTM) that can be scrambled or shuffled to non-native bonds during recombinant expression, sample handling, or sample purification. Currently, mapping of disulfide bonds is not easy because of various sample requirements and data analysis difficulties. One step towards facilitating this difficult work is developing a better understanding of how disulfide-bonded peptides fragment during collision induced dissociation (CID). Most automated analysis algorithms function based on the assumption that the preponderance of product ions observed during the dissociation of disulfide-bonded peptides result from the cleavage of just one peptide bond, and in this report we tested that assumption by extensively analyzing the product ions generated when several disulfide-bonded peptides are subjected to CID on a quadrupole time of flight (QTOF) instrument. We found that one of the most common types of product ions generated resulted from two peptide bond cleavages, or a double cleavage. We found that for several of the disulfide-bonded peptides analyzed, the number of double cleavage product ions outnumbered those of single cleavages. The influence of charge state and precursor ion size was investigated, to determine if those parameters dictated the amount of double cleavage product ions formed. It was found in this sample set that no strong correlation existed between the charge state or peptide size and the portion of product ions assigned as double cleavages. These data show that these ions could account for many of the product ions detected in CID data of disulfide bonded peptides. We also showed the utility of double cleavage product ions on a peptide with multiple cysteines present. Double cleavage products were able to fully characterize the bonding pattern of each cysteine where typical single b/y cleavage products could not.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Verification of protein three-dimensional structure is often complicated, yet necessary for both research purposes and industrial production. Disulfide bonding is one very important protein feature that can affect protein structure and functionality [1–6]. The ability of two amino acid residues in close proximity to undergo an oxidation to form a bond is unique to cysteine and has multiple spatial requirements [7]. These intra- and possibly inter-protein bridges play a large part in stabilizing the overall structure of a protein [1, 3–6]. Anfinsen et al. showed that in the absence of native disulfide bonds, some proteins cannot achieve the correct three-dimensional structure to perform their proper function [1, 5]. The determination of the disulfide bonding network of a protein is therefore one important tool for characterizing isolated and recombinantly-expressed proteins, and techniques to facilitate disulfide bonding analysis are presented herein.
There are several ways to go about determining the disulfide bonding network of a protein with mass spectrometry. Many groups have developed methods to break the disulfide bond, generating characteristic mass shifts or a reporter ion to monitor [8–14]. However, this approach is complicated because the sample preparation is somewhat complex, often requiring the addition of reagents that will cause either a characteristic mass shift (such as isotope labeling) or scission of the disulfide bond [10, 15–22]. Electron capture dissociation and electron transfer dissociation have also shown promise to potentially map disulfide bond networks through a top down approach [23, 24] as well as a bottom up approach [25]. A simpler work flow involves subjecting tryptic peptides to CID and using de novo sequencing to assign a peptide sequence [26]. However, the key limitation of this approach with disulfide-bonded peptides is data analysis. Identification of MSn data for disulfide-bonded peptides is tedious, time-consuming, and difficult-especially when more than two peptides are involved. As a result, computer algorithms are relied upon heavily to aid in the analysis.
Currently, there are several programs that assign MSn data for disulfide-bonded peptides [27–31]. Four of the most notable programs are xComb [28], MS2Assign [29], SearchXLinks [30], and MassMatrix [31]. All of these programs attempt to match product ions in the MS/MS data to a theoretical spectrum of product ions from candidate disulfide-bonded peptide sequences. Each program has its merits and limitations; however, they each share a potential weakness: in all the programs, the assumption is made that the most common fragmentation observed during CID will be the cleavage of one bond (typically a peptide bond). This assumption is likely based on the early observations from Schilling et al., who reported that the abundance of double cleavage ions was less than 10% with a quadrupole time of flight (QTOF) instrument [29]. (The terminology “double cleavage” is adopted from references [28] and [30].) Studies using electrospray ionization (ESI) have found that the number and type of cleavages can be dependent on charge [24, 25, 32–34]. If, in fact, disulfide bonded peptide ions produced during ESI typically undergo multiple bond cleavages under CID conditions, software that attempts to assign disulfide bonded peptides could be optimized by incorporating this information.
The present investigation was carried out to provide additional insight into the fragmentation trends of disulfide linked peptides, so that this fundamental knowledge can provide guidance to developers of disulfide mapping software. The key question asked here is: When can one assume that disulfide linked peptides predominantly undergo just a single peptide cleavage, after ionization by ESI-MS and CID? To answer this question, a variety of disulfide-bonded peptides are subjected to CID in an ESI-QTOF mass analyzer, and a diligent attempt is made to identify each of the product ions by considering many possible fragmentation pathways that the peptides could undergo. The product ions are assigned manually, using a rigorous set of rules for verifying the assignment. These studies ultimately indicate that cleavage of two peptide bonds is an important and common fragmentation pathway for disulfide-bonded peptides undergoing CID.
2 Experimental
2.1 Materials and Reagents
Lysozyme from chicken egg white, acetic acid, and formic acid were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ammonium bicarbonate was purchased from Fluka (Milwaukee, WI, USA). Ammonium carbonate and HPLC grade acetonitrile were purchased from Fisher (Pittsburgh, PA, USA). Sequencing-grade modified trypsin was purchased from Promega (Madison, WI, USA). The 4-vinylpyridine used was purchased from Acros Organics (Morris Plains, NJ, USA). Water was purified by a Millipore Direct-Q3 Water Purification System (Billerica, MA, USA).
2.2 Sample Preparation and Proteolysis
Samples of ~300 μg of protein were dissolved in two different ammonium carbonate 15 mM buffers, one at pH 6.0 (H2CO3/NH4HCO3) and the other at pH 9.0 (NH4HCO3/(NH4)2CO3), to approximately 3.0 μg/μL. Both were incubated at 37 °C for 1 h. Free cysteines were alkylated by the subsequent addition of 4-vinylpyridine and allowed to incubate at room temperature for 1 h in the dark. Two trypsin additions were made, each being 1:15 enzyme:substrate, 8 h apart. After 24 h from the first trypsin addition, 1 μL concentrated acetic acid per 20 μL of solution was added. Samples were analyzed immediately following acetic acid addition.
2.3 QTOF LC-MS
Liquid chromatography was performed on a Waters Acquity UPLC using a Micro-Tech Scientific capillary column (500 μm i.d., 10 cm, 3 μm, 300 Å) (Vista CA) equipped with a 5 μL injection loop. A flow rate of 5 μL/min and a linear gradient beginning at 98%:2% A:B with increasing organic concentration over 50 min was used. Solvent A consisted of 99.9%/0.08% H2O/formic acid and Solvent B consisted of 99.9%/0.08% acetonitrile/formic acid. Positive ion mode was used, and the ESI needle was held at 3.5 kV. MassLynx software was set up to perform MS2 on selected m/z ratios with a Q-TOF-2 (Micromass Ltd., Manchester, UK). The collision energy varied, depending on the m/z of the precursor ion. Between m/z 350 and 600, 25% normalized collision energy was used; between m/z 601 and 900, 30% was used; and between m/z 901 and 2000, 35% normalized collision energy was used. In the case of Peptides D, E, and F these collision energies did not adequately dissociate the precursor ion, so an additional 5% normalized collision energy was added, which yielded sufficient fragmentation.
2.4 Data Analysis
A “look-up” table of possible fragment ions was prepared for each peptide. (All look-up tables used are provided in Supplementary Material). The table contained the m/z and compositions of all possible b and y ions, ions resulting from disulfide bond scission, and ions resulting from the combination of any two single cleavages (including 2 b ions, 2 y ions, all possible combinations of 2 b/y ions, ions containing a single b or y cleavage and a disulfide scission, and any of the above single cleavage ions accompanied by loss of water or loss of NH3). Only monoisotopic masses were used in the table and in peak assignment. For each MS2 spectrum of disulfide bonded peptides, the 25 to 40 monoisotopic peaks of highest relative abundance were assigned by matching the monoisotopic mass to the masses in the custom-prepared look-up table. If the monoisotopic peak could not be clearly identified, the peak was discarded. If the ions did not match any mass in the look-up table resulting from combinations of b or y ion cleavage and/or disulfide cleavage, neutral losses of water or NH3 in combination with b or y ions were considered as a possible fragmentation pathway. (Assignment of any neutral losses required the necessary residues or termini capable of producing those neutral losses present in that peptide fragment.) Finally, for ions that remained unassigned, a and x ions were also considered as potential product ions. For MS2 data, the assigned composition for each product ion was further verified by assuring that the charge state for the experimental data matched the charge state for the ion’s assigned composition. Peaks were required to have no more than a ±0.15 Th mass error to be verified as “correctly assigned.” Peaks in spectra are labeled either as “1”, “2”, “U”, “M-H2O/M-NH3”, or “A”. These designations refer to single cleavage (1), double cleavage (2), unknown (U), precursor ion–water or ammonia (M–H2O/M–NH3), or ambiguous (A), respectively. An ambiguous ion was an ion whose m/z (and charge state) was within the mass tolerance of both a possible single and double cleavage product ion on the look-up table for that particular peptide.
3 Results and Discussion
To develop a better fundamental understanding of fragmentation of multiply-charged disulfide-linked peptides undergoing CID, a variety of analytes were generated by tryptic digestion of the protein lysozyme. The disulfide-linked peptides investigated in this study are shown in Table 1. Of these, A, B, and D are native disulfide linkages in lysozyme, and C was generated through intentional disulfide scrambling, by adjusting the pH of the solution to 9.0, prior to digestion. In addition, the fragmentation trends of two of these peptides (C and D) were assessed in various charge states. Using this data set, the dissociation characteristics of the peptides are described below, and the effects of peptide size and peptide charge state on the propensity of the peptide ion to undergo multiple b/y cleavages are determined.
3.1 Small Peptides
Do the fragmentation characteristics of disulfide-bonded peptides depend on the overall size of the peptide? To investigate this question, the fragmentation characteristics of two small disulfide-bonded peptides (Table 1A and B) containing one inter-peptide bond were investigated. Both contained a large α peptide and a much smaller β peptide. Both species were subjected to CID in a QTOF mass spectrometer in the 2+ charge state (Figure 1). After assigning all of the most abundant product ions for these two species by matching m/z and charge state of the observed product ions to an exhaustive list of theoretical product ions (as described in the Experimental section), it was apparent that the size of the peptide did not dictate the amount of single cleavage versus double cleavage ions observed (Table 2). Peptide A produced all single cleavage product ions while Peptide B produced a mix of both single cleavage and double cleavage product ions. Approximately one-third of the ions assigned for Peptide B were the result of double peptide backbone cleavages. These data demonstrate that even though the relative size of α and β peptides can be very similar, the propensity to form double cleavages is very different. Table 2 shows that if double cleavages were not included, only about half the peaks that could have been assigned in Peptide B would have been assigned.
QTOF MS2 spectra for Peptides A and B in the 2+ charge state, at m/z (a) 584 and m/z (b) 758. Each “1” refers to a single cleavage, “2” refers to a double cleavage, “U” refers to an unknown ion, and “A” refers to an ambiguous assignment. Some assignment labels have been omitted due to space constraints
3.2 Medium Peptide
After investigating the fragmentation patterns of two small disulfide-bonded peptides, the next step in our comprehensive fragmentation analysis of these types of ions was to contrast the fragmentation pattern of small disulfide-bonded peptides to medium and large species. To generate a “medium” sized peptide, a non-native lysozyme disulfide-bonded peptide containing one inter-peptide bond was formed, through pH and temperature control. This peptide is labeled C in Table 1, and it exemplifies a peptide that contains a medium-sized peptide for both the α and β chains. When fragmented in two different charge states (2+ and 3+), Peptide C formed a mix of single and double cleavage product ions (Table 2). One-fifth of the ions assigned in the higher charge state were double cleavages and approximately one-half of the ions assigned in the lower charge state were double cleavages. Comparing Peptides B and C, one can see that even though they have quite different compositions, concerning α and β peptide relative size, they fragment in similar quantities of single and double cleavage product ions. These data support the conclusion that the mass of the α/β chains cannot be used to predict whether or not the peptide will undergo multiple cleavages during CID.
3.3 Large Peptide
The fragmentation of a “large peptide” was also investigated for these studies. Tryptic digestion of lysozyme yielded one disulfide-bonded peptide containing one intra-peptide bond as well as one inter-peptide bond. This peptide is labeled D, shown in Table 1, and the MS/MS data for this peptide (in two different charge states) is shown in Figure 2a and b. When subjected to CID, the peptide formed numerous double cleavages and few single cleavage product ions (Table 2). This peptide had the largest proportion of peaks assigned as double cleavages of the four analyzed. The product ions for this species were assigned from precursors of two different charge states. Both charge states analyzed (3+ and 4+) showed a large proportion of ions resulting from cleavage of at least two bonds. However, many more high-abundance peaks were assigned as double cleavages in the higher charge state.
(a) QTOF MS2 spectrum for Peptide D in the 4+ charge state at m/z 817. Each “1” refers to a single cleavage and “2” refers to a double cleavage. (b) QTOF MS2 spectrum for Peptide D at m/z 1090, an ion in the 3+ charge state. Each “1” refers to a single cleavage, “2” refers to a double cleavage, “U” refers to an unknown ion, and “A” refers to an ambiguous assignment. Some assignment labels have been omitted due to space constraints
3.4 Effect of Charge State
Peptides C and D gave a sufficient signal in the CID spectra to warrant analysis in multiple charge states, as summarized in Table 2. These two peptides provided conflicting data in the attempt to find a way to associate relative charge state with the amount of ions formed as a result of multiple bond cleavages. Peptide C was analyzed in both the 2+ and 3+ charge states. The lower charge state had several more peak assignments that consisted of double cleavage products. Peptide D was analyzed in both the 3+ and 4+ charge states. However, this example showed the higher charge state to have a much higher population of double cleavage product ions. While this data set is small, it is large enough to indicate that there is no strong correlation between the charge state and the type of ions formed.
3.5 Double Cleavages are Significant, but Cannot be Predicted
The presence of double cleavage product ions in low energy collision induced dissociation spectra has been noticed previously, but the general perception is that these ions are typically in relatively low abundance [27, 29]. Our results show that the formation of double cleavage product ions in disulfide-bonded peptide CID spectra is significantly more abundant than previously reported. As can be seen with the Peptides C and D, the majority of the high abundance peaks can consist of double peptide backbone cleavages. Likewise, extensive fragmentation was also observed for a small disulfide-bonded peptide, and the charge state of the ion did not appear to dictate the propensity for these ions to form. Since these ions cannot be suppressed or readily predicted to be present or absent, one should henceforth consider the likely possibility that a significant number of product ions could be from the cleavage of at least two bonds when an unknown sample is subjected to CID for the purpose of disulfide mapping. The addition of these possible fragmentation pathways in peak assignment algorithms will allow for a more thorough analysis of peaks and may generate more confident peptide assignments; however including these additional ions may not guarantee a lower false-positive discovery rate.
3.6 Validation
In an effort to validate the above findings, another data set that was 50% the size of the original set was analyzed. A peptide with an intrapeptide disulfide bond from bovine serum albumin was fragmented in one charge state (labeled Peptide E) and a peptide with an interpeptide disulfide bond from bovine fetuin was fragmented in two different charge states (labeled Peptide F), See Tables 1 and 2. The data (included in Supplementary Material and Table 2) show that, again, the number of double cleavage product ions can be as plentiful if not more so than single cleavage products, for various types of disulfide-linked peptides.
Additionally, a recent analysis of the disulfide linkage pattern of a recombinant HIV-Env protein used CID data to assign disulfide bonds [35], and the study further supports our findings that many double cleavage product ions are generated upon CID of disulfide linked peptides. A total of 12 different disulfide-linked peptides of a variety of sizes and disulfide-linkage types were characterized using CID data in the 2+, 3+, and 4+ charge states. About 340 product ions were assigned in the MS/MS spectra. Of those product ions, about 48% (164 ions) were the result of cleavage of at least two peptide bonds, (see Supplemental Data in reference [35]). This study further validates the findings described here, that double cleavage ions can be abundant, regardless of the peptide size or charge state.
3.7 Utility of Double Cleavages for Disulfide Bond Elucidation
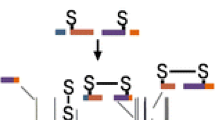
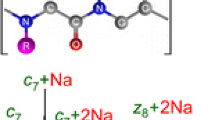
Double cleavage product ions can provide valuable insight into the disulfide network, even when multiple cysteine residues reside on a single peptide. (As an example, see Peptide D in Table 1.) The presence of more than two cysteines in a disulfide-linked peptide complicates disulfide analysis because even if one knows which peptides are bonded to each other, the disulfide connectivity is impossible to infer. In Peptide D in Table 1, three cysteines are present in one tryptic peptide, and this peptide is disulfide-bonded to a peptide containing one cysteine. This particular peptide presents an interesting case, since single cleavage product ions will not provide significant information about the disulfide networking due to the intra-peptide bond. Double cleavage assignments were useful in verifying the bonding of each separate cysteine residue. The MS/MS data for this peptide is in Figure 2a and b, and these data show a double cleavage peak at 904.33 Th. (This ion appeared in both the 3+ and 4+ charge states.) The only possibility for this mass and charge state is that of a double cleavage ion involving only the center of the three cysteine residues and the other peptide to which it is disulfide-bonded, as shown in Figure 3. Thus, using this ion allowed for the determination of the bonding structure of all the cysteines: Two must be intra-peptide bonded and the center is bonded to another peptide. If double cleavages had been ignored, full characterization of the bonding structure of the peptide would have been impossible, without further studies.
4 Conclusion
Acquiring a better fundamental understanding of disulfide-linked peptides’ fragmentation is an important first step in developing better automated approaches to assigning disulfide bonding using MS/MS data. We contribute to this field by asking the question: How prevalent are multiple bond cleavages of disulfide-linked peptides, when the species undergo collision induced dissociation? After thoroughly analyzing MS/MS data for several different types of disulfide linked peptides, we unequivocally showed that product ions resulting from cleavage of at least two peptide backbone bonds (double cleavages) are prevalent in many different disulfide linked peptides’ MS/MS data. Furthermore, the precise amount of double cleavages cannot be linked to charge state or the size of the peptide. This information on the abundant presence of double cleavages in MS/MS analysis of disulfide-linked peptides is important because it could be incorporated into disulfide analysis algorithms to increase the number of product ions assigned in the MS/MS data. Additionally, we showed that assigning double cleavage ions is useful when peptides contain more than two cysteines, because these product ions can provide information about the connectivity of the cysteines.
References
Anfinsen, C.B.: Principles that govern the folding of protein chains. Science 181, 223–230 (1973)
Arnesano, F., Banci, L., Bertini, I., Martinelli, M., Furukawa, Y., O'Halloran, T.V.: The unusually stable quaternary structure of human Cu, Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 279, 47998–48003 (2004)
Yano, H., Kuroda, S., Buchanan, B.B.: Disulfide proteome in the analysis of protein function and structure. Proteomics 2, 1090–1096 (2002)
Angata, K., Yen, T.Y., El-Battari, A., Macher, B.A., Fukuda, M.: Unique disulfide bond structures found in ST8Sia IV polysialyltransferase are required for its activity. J. Biol. Chem. 276, 15369–15377 (2001)
Anfinsen, C.B., Haber, E.: Studies on the reduction and re-formation of protein disulfide bonds. J. Biol. Chem. 236, 1361–1363 (1961)
Givol, D., De Lorenzo, F., Goldberger, R.F., Anfinsen, C.B.: Disulfide interchange and the three-dimensional structure of proteins. Proc. Natl. Acad. Sci. U. S. A. 53, 676–684 (1965)
Creighton, T.E.: Disulfide, bond formation in proteins. Methods Enzymol. 107, 305–329 (1984)
Kim, H.I., Beauchamp, J.L.: Mapping disulfide bonds in insulin with the route 66 method: selective cleavage of S–C bonds using alkali and alkaline earth metal enolate complexes. J. Am. Soc. Mass Spectrom. 20, 157–166 (2009)
King, G.J., Jones, A., Kobe, B., Huber, T., Mouradov, D., Hume, D.A., Ross, I.L.: Identification of disulfide-containing chemical cross-links in proteins using MALDI-TOF/TOF-mass spectrometry. Anal. Chem. 80, 5036–5043 (2008)
Sinz, A.: Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 38, 1225–1237 (2003)
Soderblom, E.J., Bobay, B.G., Cavanagh, J., Goshe, M.B.: Tandem mass spectrometry acquisition approaches to enhance identification of protein–protein interactions using low-energy collision-induced dissociative chemical crosslinking reagents. Rapid Commun. Mass Spectrom. 21, 3395–3408 (2007)
Soderblom, E.J., Goshe, M.B.: Collision-induced dissociative chemical cross-linking reagents and methodology: applications to protein structural characterization using tandem mass spectrometry analysis. Anal. Chem. 78, 8059–8068 (2006)
Thakur, S.S., Balaram, P.: Fragmentation of peptide disulfides under conditions of negative ion mass spectrometry: studies of oxidized glutathione and contryphan. J. Am. Soc. Mass Spectrom. 19, 358–366 (2008)
Chrisman, P.A., McLuckey, S.A.: Dissociations of disulfide-linked gaseous polypeptide/protein anions: ion chemistry with implications for protein identification and characterization. J. Proteome Res. 1, 549–557 (2002)
Pearson, K.M., Pannell, L.K., Fales, H.M.: Intramolecular cross-linking experiments on cytochrome c and ribonuclease a using an isotope multiplet method. Rapid Commun. Mass Spectrom. 16, 149–159 (2002)
Yen, T.Y., Macher, B.A.: Determination of glycosylation sites and disulfide bond structures using LC/ESI-MS/MS analysis. Methods Enzymol. 415, 103–113 (2006)
Yen, T.Y., Yan, H., Macher, B.A.: Characterizing closely spaced, complex disulfide bond patterns in peptides and proteins by liquid chromatography/electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 37, 15–30 (2002)
Rinner, O., Seebacher, J., Walzthoeni, T., Mueller, L., Beck, M., Schmidt, A., Mueller, M., Aebersold, R.: Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 (2008)
Mann, M., Jensen, O.N.: Proteomic Analysis of Post-Translational Modifications. Nat. Biotechnol. 21, 255–261 (2003)
Qin, J., Chait, B.T.: Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Anal. Chem. 69, 4002–4009 (1997)
Yen, T.Y., Joshi, R.K., Yan, H., Seto, N.O.L., Palcic, M.M., Macher, B.A.: Characterization of cysteine residues and disulfide bonds in proteins by liquid chromatography/electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 35, 990–1002 (2000)
Mentinova, M., Han, H., McLuckey, S.A.: Dissociation of disulfide-intact somatostatin ions: the roles of ion type and dissociation method. Rapid Commun. Mass Spectrom. 23, 2647–2655 (2009)
Ge, Y., Lawhorn, B.G., ElNaggar, M., Strauss, E., Park, J.-H., Begley, T.P., McLafferty, F.W.: Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J. Am. Chem. Soc. 124, 672–678 (2002)
Liu, J., Gunawardena, H.P., Huang, T.Y., McLuckey, S.A.: Charge-dependent dissociation of insulin cations via ion/ion electron transfer. Int. J. Mass Spectrom. 276, 160–170 (2008)
Gunawardena, H.P., Gorenstein, L., Erickson, D.E., Xia, Y., McLuckey, S.A.: Electron transfer dissociation of multiply protonated and fixed charge disulfide linked polypeptides. Int. J. Mass Spectrom. 265, 130–138 (2007)
Standing, K.G.: Peptide and protein de novo sequencing by mass spectrometry. Curr. Opin. Struct. Biol. 13, 595–601 (2003)
Gao, Q.X., Xue, S., Doneanu, C.E., Shaffer, S.A., Goodlett, D.R., Nelson, S.D.: Pro-CrossLink. Software tool for protein cross-linking and mass spectrometry. Anal. Chem. 78, 2145–2149 (2006)
Panchaud, A., Singh, P., Shaffer, S.A., Goodlett, D.R.: xComb: a cross-linked peptide database approach to protein–protein interaction analysis. J. Proteome Res. 9, 2508–2515 (2010)
Schilling, B., Row, R.H., Gibson, B.W., Guo, X., Young, M.M.: MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 (2003)
Wefing, S., Schnaible, V., Hoffmann, D.: SearchXLinks. A program for the identification of disulfide bonds in proteins from mass spectra. Anal. Chem. 78, 1235–1241 (2006)
Xu, H., Zhang, L.W., Freitas, M.A.: Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J. Proteome Res. 7, 138–144 (2008)
Wells, J.M., Stephenson, J.L., McLuckey, S.A.: Charge dependence of protonated insulin decompositions. Int. J. Mass Spectrom. 203, A1–A9 (2000)
Hogan, J.M., McLuckey, S.A.: Charge state dependent collision-induced dissociation of native and reduced porcine elastase. J. Mass Spectrom. 38, 245–256 (2003)
Lioe, H., O'Hair, R.A.J.: A novel salt bridge mechanism highlights the need for nonmobile proton conditions to promote disulfide bond cleavage in protonated peptides under low-energy collisional activation. J. Am. Soc. Mass Spectrom. 18, 1109–1123 (2007)
Go, E. P.; Zhang, Y.; Menon, S.; Desaire, H.; Analysis of the disulfide bond arrangement of the HIV envelope protein CON-S gp140 ∆CFI shows variability in the V1 and V2 regions. J. Proteome Res. (2011), In Press.
Acknowledgments
The authors acknowledge support for this work by NIH grant RO1GM077226. Daniel Clark also acknowledges support from an NIH training grant, number T32 GM008545. They thank the Mass Spectrometry Laboratory at KU for instrument time.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Material 1
(XLS 294 kb)
Rights and permissions
About this article
Cite this article
Clark, D.F., Go, E.P., Toumi, M.L. et al. Collision Induced Dissociation Products of Disulfide-Bonded Peptides: Ions Result from the Cleavage of More Than One Bond. J. Am. Soc. Mass Spectrom. 22, 492–498 (2011). https://doi.org/10.1007/s13361-010-0064-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-010-0064-x