Abstract
This study analysed the phenotypic and genotypic variation among 511 Pseudomonas savastanoi pv. phaseolicola (Psp) isolates, causing halo blight in mungbeans. Collected from symptomatic mungbean (Vigna radiata) crops throughout Australia between 2005 and 2018, a total of 352 Psp isolates were phenotypically screened. Our in planta screening against a set of four mungbean cultivars with known susceptible and resistant reactions revealed five distinctive pathotypes. Isolates belonging to pathotype 2 were the most prevalent at 84% and were found to be highly pathogenic towards all tested mungbean genotypes. Genomic variation was investigated for 205 isolates using DNA fingerprints, splitting the halo blight pathogen population into two broad genetic lineages. Further genetic testing for two known avirulence genes, avrPphE and avrPphF, identified the avrPphE gene in all the tested isolates and avrPphF present in all but two. To identify candidate avirulence genes unique to Psp isolates infecting mungbean in Australia, a comparative genomics study was undertaken on the whole-genome sequences of two epidemiologically important Psp isolates, T11544 and K4287. The information presented in this study has the potential to dramatically improve mungbean disease resistance now and into the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mungbean (Vigna radiata L. Wilczek var. radiata) is a grain legume that provides a vital source of nutrition for many countries and contributes significantly to Australian agricultural exports (Noble et al. 2019; Shanmugasundaram et al. 2009). Severely limiting the production of commercial mungbean crops in Australia is the seed-borne bacterial disease halo blight, caused by Pseudomonas savastanoi pv. phaseolicola. An emerging threat in 1980, halo blight is now responsible for large-scale annual losses in mungbean crops and threatens the sustainability of the industry (Noble et al. 2019). Knowledge of genetic variation within the pathogen population and pathogenicity towards elite germplasm is crucial to developing multifaceted management options that will secure the future sustainability of mungbean production.
Taylor et al. (1996a), used a global study of 859 isolates from 303 disease occurrences from the common bean differential set to assign nine races of P. savastanoi pv. phaseolicola. Arnold et al. (2011) further refined the P. savastanoi pv. phaseolicola race structure by calculating the frequency of resistance (R) and avirulence (avr) genes among the host race typing set and global pathogen population, finding that a higher R gene frequency in the host led to a lower matching avr gene frequency in the pathogen population. This suggests that when an R gene is extensively deployed, races of the pathogen carrying the corresponding avr gene are suppressed (Arnold et al. 2011). The common bean differential set has since been used to assess the race diversity of 30 P. savastanoi pv. phaseolicola strains isolated from Australian mungbeans. Race 7, as described by Taylor et al. (1996b), was identified as well as variants that did not coincide with the race structure reported for common bean (Ryley et al. 2010; Taylor et al. 1996b).
This study characterises 511 P. savastanoi pv. phaseolicola isolates sampled from the naturally occurring population in Eastern Australia, focusing primarily on the South East Queensland region. In doing so, the isolates were categorised into five pathotypes and two broad genetic linages. Two isolates with markedly different pathologies were further investigated using whole genome sequences to compare virulence factors. The findings presented here will improve basic research and applied outcomes for mungbean researchers, breeders and industry.
Materials and methods
P. savastanoi pv. phaseolicola bacterial strains isolated from Vigna radiata
A total of 511 P. savastanoi pv. phaseolicola isolates were isolated from symptomatic mungbean leaf tissue, stored at either the Queensland Department of Agriculture and Fisheries (DAF) or the Queensland University of Technology (QUT) (Table S1). Symptomatic leaves were surface-sterilized by spraying with 70% ethanol and rubbing with sterile lint-free tissues several times. Lesions were dissected from the leaf and bisected with one piece observed for ooze using a compound microscope and the other placed in a drop of sterile water for bacterial elution. Following confirmation of oozing, a sterile loop was used to streak the water suspension onto King’s B (KB) medium (King et al. 1954). The plates were incubated at 28 °C for 24 h. Single colonies were twice sub-cultured to obtain pure cultures. A loop of the pure culture suspended in a 500 μL aliquot of sterile water and used directly as the template in molecular assays. Bacterial suspensions were stored at −20 °C, and for long-term storage, suspended duplicates of each isolate were held in 50% glycerol at −80 °C.
Host-pathogen interactions based on the phenotypic assessment of disease reactions on four Vigna radiata genotypes
In consultation with the Australian National Mungbean Breeding Improvement Program (NMIP), four host genotypes of mungbean (V. radiata) were selected based on disease reactions observed in artificially inoculated glasshouse and field disease screening experiments. Two seeds of each member of the differential set were sown into commercial potting mix (Rocky Point Mulching™) in seedling trays consisting of 42 wells 4 cm3 in volume. For each genotype, six wells were used and thinned to one seedling per well after emergence. Seedlings were maintained in a growth cabinet at 22± 5 °C under a 12 h light/12 h dark regime and watered regularly. Before the emergence of the first leaves, pure cultures of each isolate were spread on KB medium and incubated at 28 °C for 72 h. A sterile needle dipped into a single colony was pierced through the leaves. As a control treatment, six seedlings per tray were inoculated with a sterile needle dipped in sterile water. After inoculation, seedlings were sprayed briefly with a handheld spray bottle containing sterile distilled water and covered with a plastic bag. After 48 h the plastic bags were removed. Inoculated leaves were assessed for the presence of a chlorotic halo at the point of inoculation and small, circular, dark brown water-soaked lesions fourteen days after inoculation. Genotypes displaying characteristic symptoms were recorded as susceptible, while those with only a necrotic lesion at the point of inoculation were considered resistant. Isolations were made from the symptomatic tissues of randomly selected genotypes using the methods outlined previously. Variation in pathogenicity was assessed for a total of 352 P. savastanoi pv. phaseolicola isolates.
Genomic profiling and amplification of avirulence genes
The presence or absence of avirulence genes and DNA fingerprints were generated for 205 isolates of P. savastanoi pv. phaseolicola (primers listed in Table S2). PCR conditions for each of the primers were as described in the literature: ERIC (Versalovic et al. 1991), BOX (Versalovic et al. 1994), IS50 (Sundin et al. 1994), avrPphE (Stevens et al. 1998) and avrPphF (Tsiamis et al. 2000). In brief, 10 μL reactions contained 1 μL of bacterial cells at a concentration of ~1 × 106 CFU mL−1 as template, the final primer concentration in the reaction was 50 pM and PCR programs were as described in each reference. Thermal cycling was conducted using an Applied Biosystems, Life Technologies Proflex PCR system and products were separated by electrophoresis on 1% agarose gels, cast and run in 0.5x TAE buffer. Bands were visualised using SybrSafe stain and the G-Box Syngene gel documentation system.
Cloning and sequence analysis of the amplicon unique to the IS50-B genotype
To identify the DNA sequence and unique variations present in the PCR product of IS50-B, the 500 bp amplicon was excised and cloned into a pGEM-T Easy vector (Promega). A total of eight sequences were obtained from three isolates T13733B, T13804A, and T14028 by Sanger sequencing (Macrogen, Seoul, Rep. of Korea) using the M13 forward primer. Sequencing results were checked using Vector NTI and BLAST analysis against the NCBI database and assembled genomes from the study of T11544 and K4287. Protein modelling of the sequence through Phyre2 considered hits with homology confidence of >99 to determine possible functions (Kelley et al. 2015).
Whole-genome comparison of two epidemiological significant P. savastanoi pv. phaseolicola isolates T11544 and K4287
Isolated from infected mungbean leaf tissue in 2005, isolate T11544 has been used to challenge germplasm and breeding lines in the Australian National Mungbean Improvement Program for over a decade. However, another strain present in Australia, K4287 isolated in 2013, is highly virulent and overcomes the defences of the germplasm used as resistance donors against the T11544 strain. Before high-throughput sequencing of P. savastanoi pv. phaseolicola strains K4287 and T11544, pathogenicity was confirmed on V. radiata. DNA was extracted from pure plate cultures of each isolate using a DNeasy Blood & Tissue Kit (QIAGEN). Library preparation and sequencing were performed by Macrogen, using the TruSeq Nano DNA Kit for library preparation (TruSeq Nano DNA Sample Preparation Guide, Part # 15041110 Rev. A) and high-throughput sequencing was conducted using an Illumina HiSeq 2500 System (User Guide Document # 15035786 v01HCS 2.2.70). In total, 26,385,408 and 27,767,198 paired-end reads were generated for T11544 and K4287, respectively.
Adapter sequences and low-quality sequences based on quality scores of Phred <30 and a minimum length of 50 bp were removed using Trim Galore (version 0.5.0) (Krueger 2015). After quality control, the Illumina reads were de novo assembled using the SPAdes (version 3.13.0) genome assembly algorithm (Bankevich et al. 2012). Completeness of the assembly was assessed using QUAST (Quality Assessment Tool for Genome Assemblies, version 5.0.2) (Gurevich et al. 2013). Assembled contigs were annotated using the RAST annotation pipeline (Aziz et al. 2008).
The de novo assembled K4287, and T11544 genomes were aligned and compared using Mauve (Darling et al. 2004). Genome-wide comparison of sequence similarity was conducted with BLASTN, filtering scaffolds with > = 50% coverage of the queried sequence. BLASTP compared protein-coding genes hits, filtering those with a coverage > = 95% and sequence similarity of > = 98%. Average Nucleotide Identity (ANI) was calculated between the K4287 and T11544 genome assemblies as described by Yoon et al. (2017).
The raw Illumina data has been deposited in the NCBI Short Read Archive database under SRA accession PRJNA603636 for the T11544 and K4287 genomes. The whole-genome shotgun (WGS) draft genome assemblies were deposited in GenBank under the accession numbers JAAFOY000000000 for P. savastanoi pv. phaseolicola T11544 strain and JAAFOZ000000000 for P. savastanoi pv. phaseolicola K4287 strain.
Results
Host-pathogen interactions reveal five pathotypes of P. savastanoi pv. phaseolicola infecting Australian mungbeans
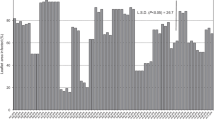
The existence of five pathotypes among 352 P. savastanoi pv. phaseolicola isolates was revealed based on the pathogenicity of the isolates towards four V. radiata genotypes (Table 1). Of the five disease reactions observed, isolates belonging to pathotype 2 were the most pathogenic, producing symptoms on all genotypes. Isolated from all growing regions throughout Australia (Fig. 1), pathotype 2 comprised 84% of all isolates screened in this study (Table 1). In contrast, isolates from pathotype 1 are only pathogenic towards the commercial cultivar Crystal and accounted for 10% of the population (Table 1). The remaining pathotypes, 3, 4 and 5 combined comprise as little as 6% of the population and exhibit cultivar-specific virulence (Table 1). Based on prior knowledge and results from this study, isolates T11544 and K4287 were selected for further investigation. Isolate K4287 represents isolates from pathotype 2, identified to overcome known resistance in glasshouse screening (Table 1). Isolate T11544, used as the primary source of inoculum in resistance breeding, represents pathotype 1 as a direct comparison.
Genomic profiling of the P. savastanoi pv. phaseolicola population in Australia
Molecular markers reveal the population of Australian P. savastanoi pv. phaseolicola is highly conserved (Fig. 2) and splits into two broad genetic lineages (Fig. 3). The ERIC and BOX primers, previously used in genomic profiling to distinguish between bacterial strains within P. savastanoi pathovars (Weingart and Völksch 1997), revealed no differences among representatives from the five pathotypes analysed here. In contrast, the IS50-PCR DNA fingerprints distinguished two patterns, “A” and “B”, which were discernible by the presence or absence of a specific amplicon of approximately 500 bp (Fig. 3). A subset of 58 isolates from 2005 to 2016 were primarily categorised as sub-populations IS50-A (75%) and IS50-B (25%). In comparison, the 148 P. savastanoi pv. phaseolicola isolates from 2017 and 2018 revealed a pronounced decrease in IS50-A to 55%, while IS50-B increased to 45% of all isolates (Fig. 4). This difference is statistically significant at the 1% confidence level (chi-square statistic with Yates correction = 6.9388, p value = 0.008435). Sub-population IS50-A concentrated in South East Queensland was revealed as decreasing in size over time, while sub-population IS50-B grew by 80% in 2017 and 2018, spreading north to Central Queensland and south to Northern New South Wales. Isolates T11544 and K4287 were categorised as IS50-A, suggesting other factors are affecting virulence.
Examples of DNA fingerprints from P. savastanoi pv. phaseolicola using IS50 PCR primers. Lane 01: 2-Log DNA Ladder (0.1–10.0 kb) (New England Biolabs); Lanes 02: T11544; 03: T14006; 04: K4287; 05: T14031; 06: T13913; 07: T13976; 08: T13994; 09: T13733B;10: T13804A; 11: T13805H; 12: T13992; 13: T13754B;14: T13734A; 15: T13804F; 16: T13982; 17: T14007; 18: T13636C; 19: T13981
A total of 205 P. savastanoi pv. phaseolicola isolates were geotagged and screened with IS50 PCR primers, dividing the population into two broad genotypes. The panel is a map of mungbean growing districts in Australia highlighting the distribution of the isolates throughout Southern and Central Queensland and Northern New South Wales. The two IS50 groups are designated red for IS50-A and blue for IS50-B
The ~500 bp fragment cloned from IS50-B showed a 99.5% identity to a hypothetical protein encoded by “Pseudomonas syringae pv. cerasicola strain CFBP 6110 genome assembly, plasmid: PP2”. BLASTn returned no significant hits when using the K4287 and T115544 genome assemblies from this study as custom databases. Further to this, BLASTx did not return any hits or identify conserved domains. Analysis of the sequence using Phyre2 modelling software revealed multiple significant hits for kinase-related proteins.
Presence/absence of cloned avirulence genes avrPphE and avrPphF
The avrPphE gene, cloned from race 4 but present in all races, is reported to affect cultivar-specific avirulence in common bean (Mansfield et al. 1994; Nimchuk et al. 2007; Stevens et al. 1998). The avrPphF gene, located within a pathogenicity island on the plasmid (pAV511) was cloned from race 5 and 7 of P. savastanoi pv. phaseolicola (Tsiamis et al. 2000). Of the 205 isolates analysed in this study, all carry avirulence gene avrPphE. Two isolates PSP023 (Fig. 4) and T13733B (pathotype 2), were identified as missing avrPphF but otherwise had the same genotypic grouping IS50-B (Table S1).
Whole-genome comparison of two epidemiological significant strains of P. savastanoi pv. phaseolicola T11544 and K4287
To identify candidate virulence genes unique to strains endemic to Australia, the whole-genome sequences of strains T11544 (pathotype 1) and K4287 (pathotype 2) were assembled de novo, annotated and comparatively analysed. Both had highly similar genomes with an average nucleotide identity of 99.98% (Table S3) (Yoon et al. 2017) comprising genome sizes of ~6 Mbp (Table 2, Table S4). A Benchmarking Universal Single-Copy Orthologs’s (BUSCO) (Simão et al. 2015) analysis determined that both genome assemblies were 99.32% complete. When comparing the two genomes, 49,330 additional base pairs were identified as unique to the T11544 genome, of which ~40 kbp were located on the chromosome (Table S4). Strain K4287 had a smaller genome by approximately 50 kbp. The majority of the difference in genomic material between the strains was missing from the chromosome of K4287, while its plasmids A and B gained ~6 kbp and 4 kbp respectively (Table S4). The additional unique genetic material from both strains comprised a total of 29 annotated regions primarily associated with virulence (Table 3). Assessment of the repertoire of type III secretion proteins in both K4287 and T11544 genome assemblies identified the same 17 proteins conserved in both genomes with 100% nucleotide sequence similarity (Table S5).
Discussion
Known for over 90 years, halo blight disease of the Fabaceae family continues to threaten food production globally (Arnold et al. 2011; Burkholder 1926; Noble et al. 2019). However, limited research has explicitly focused on the interactions between the causal agent P. savastanoi pv. phaseolicola and Vigna radiata (Noble et al. 2019; Sun et al. 2017). This study provides both a broad and in-depth investigation of the phenotypic and genotypic variation that exists within the population of P. savastanoi pv. phaseolicola infecting Australian mungbeans. In addition to the identification of characterised and unique virulence factors, the categorisation of five pathotypes provides vital information to inform future breeding practices and research.
In Australia, races 2 and 7 of the nine global races were first identified from Macroptilium atropurpureum and Neonotonia wightii (Taylor et al. 1996a). In 2010, race 7 was reported as the cause of halo blight disease in Australian mungbeans with variations acknowledged that did not match any of the nine global races (Ryley et al. 2010). Four mungbean accessions with known reactions to halo blight disease were selected to dissect how those variations affect the pathogenicity towards mungbean in Australia. Of the five pathotypes described in this study, those assigned to highly pathogenic pathotype 2 accounted for 84% of all the isolates screened (Table 1). Widely dispersed across farming land, the majority of isolates screened belonged to this pathotype (Fig. 1).
Pathogenicity of bacterial pathogens is highly regulated by pairs of corresponding avirulence (avr) and resistance (R) genes in the pathogen and host, respectively (Flor 1971). Effector proteins produced by avirulence genes are injected into the host via the type three secretion systems (T3SS) found in P. savastanoi pv. phaseolicola, catalysing the infection process or inducing a hypersensitive response if a matching R gene is present (Alfano and Collmer, 1997). Isolated and sequenced from race 4, strain 1302A of P. savastanoi pv. phaseolicola, avrPphE matches the R2 gene for resistance in Phaseolus cultivars (Mansfield et al. 1994). Family members of avrPphE are prevalent among a diverse background of pathogenic bacteria suggesting a conserved role in virulence (Lindeberg et al. 2005; Nimchuk et al. 2007). Consistent with those reports, this study identified avrPphE in all the tested isolates, and avrPphF in all but two (Table S1). A loss of virulence towards susceptible P. vulgaris cultivars has been reported for strains of P. savastanoi pv. phaseolicola missing the pAV511 plasmid where avrPphF is located (Tsiamis et al. 2000). This was not the case for isolate T13733B which was missing avrPphF but retained its virulence against all four of the genotypes screened in this study. This suggests that other virulence factors are responsible for cultivar specific avirulence, or that the relevant genes have been transferred to the chromosome.
Genomic profiling further revealed a highly conserved genome at the population level (Fig. 2), delineating into two subpopulations: IS50-A and IS50-B (Fig. 3). Analysis of the amplicon unique to IS50B was limited to the identification of a hypothetical protein homolog on a plasmid from Pseudomonas syringae pv. cerasicola when interrogating the NCBI database. Protein modelling based on the sequence suggests the translated product belongs to the kinase family. The characterisation of a kinase protein linked to pathogenicity may represent an advance in understanding the mechanisms of bacterial infection. The eukaryote-like Serine Threonine Protein Kinase (STPK) family discovered in pathogenic bacteria from the genus Yersinia (Håkansson et al. 1996) was reported to play a role in sabotaging specific host defence via the G protein pathway, which is universal in plants and animals (Canova and Molle 2014; Håkansson et al. 1996). Further studies will be required to confirm the role of the uncharacterised protein, to identify if it belongs to the type III effector protein family, and whether it can be translocated to plant cells.
The majority (84%) of isolates categorised as pathotype 2 infected all four of the mungbean genotypes tested (Table 1). Further to this, all isolates contained the avrPphE gene, and all but two amplified the avrPphF gene (Fig. 5, Table S1). This suggests the matching R genes are absent from the four V. radiata genotypes tested. If for example, commercial mungbean varieties contained the R genes associated with avrPphE or avrPphF, it would be expected that strains carrying those genes would be suppressed (Arnold et al. 2011; Taylor et al. 1996a). Thus, it is likely that current varieties are particularly vulnerable to halo blight disease because they do not contain the gene-for-gene resistance that results from interactions between avr and R genes. Celera II–AU, released in 2015, is an exception and represents the gold standard for resistance to halo blight disease in Australian commercial mungbeans. However, its small seed makes it suitable for a limited number of markets, so it accounts for less than 10% of total Australian production (AMA 2015). Released in 2013, Jade-AU gained a 12% yield advantage over its predecessor Crystal but does not contain the resistance found in Celera II-AU (AMA 2015). The wide-release and adoption of new varieties bred for resistance against T11544 represents a selection pressure that has likely influenced the population of P. savastanoi pv. phaseolicola (Table 1, Fig. 4). Therefore, research, breeding and industry practices will need to adapt to stay ahead of the evolution of new pathogenic strains.
The identification of avrPphE and avrPphF throughout the pathogen population provides new targets that could significantly decrease the incidence of halo blight disease. Primary resistance could be integrated into commercial mungbean through the introgression of the R genes associated with avrPphE (Stevens et al. 1998) and avrPphF (Tsiamis et al. 2000). Screening and analysis of large germplasm resources have identified dominant resistance genes and assisted in the efficient integration of disease resistance in Phaseolus vulgaris (Tock et al. 2017). Large germplasm sets representing the global diversity of V. radiata have been sequenced and characterised to conduct similar studies investigating V. radiata disease resistance (Breria et al. 2019; Noble et al. 2017). Monitoring the pathogen population as it adapts to the release of major R gene resistance will be required as gene-for-gene resistance can break down. To preserve the longevity of newly deployed resistance, rigid integrated disease management practices, must be adhered to, such as seed screening, phytosanitation and rotation of crops.
Comparative analysis between P. savastanoi pv. phaseolicola and P. savastanoi pv. tomato DC3000 has previously revealed a high degree of conservation at the gene and genome level, with variation among genes involved in virulence and transposition (Joardar et al. 2005). A near perfect sequence similarity was observed (99.98%) in a fraction of 84% of the whole-genome sequences with orthologous relationships between isolates T11544 and K4287 from pathotypes 1 and 2 (Supplementary Table S4). Both genomes were ~ 6 Mbp and highly conserved except the less virulent T11544 strain which contained an extra ~50 kbp in sequence primarily related to virulence proteins. The coding sequences were located close together on three contigs T11544_000105 and T11544_000074 and T11544_000128 (Table 3).
Commercial varieties and breeding material have been selected for their defences against the virulence proteins found in T11544 due to its use as inoculum for field resistance screening. This has potentially allowed a highly virulent population of P. savastanoi pv. phaseolicola to thrive unknown until now (Table 1). The information presented here provides clear direction and targets for breeders and researchers to reduce the risk of halo blight on mungbean crops.
References
Alfano JR, Collmer A (1997) The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J Bacteriol 179:5655–5662
AMA (2015) Australian Mungbean Association About Us. Australian Mungbean Association (AMA). http://www.mungbean.org.au/about-us.html#industry-size-and-value. Accessed 15/10/2018
Arnold DL, Lovell HC, Jackson RW, Mansfield JW (2011) Pseudomonas syringae pv. phaseolicola: from ‘has bean’ to supermodel. Mol Plant Pathol 12:617–627. https://doi.org/10.1111/j.1364-3703.2010.00697.x
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Breria CM et al (2019) Population structure of the world vegetable center mungbean mini core collection and genome-wide association mapping of loci associated with variation of seed coat luster. Trop Plant Biol:1–12. https://doi.org/10.1007/s12042-019-09236-0
Burkholder WH (1926) A new bacterial disease of the bean. Phytopathology 16:915–927
Canova MJ, Molle V (2014) Bacterial Serine/Threonine Protein Kinases in Host-Pathogen Interactions. 289:9473–9479. https://doi.org/10.1074/jbc.r113.529917
Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. https://doi.org/10.1101/gr.2289704
Flor HH (1971) Current status of the gene-for-gene concept. Ann Rev Phytopathol 9:275–296. https://doi.org/10.1146/ANNUREV.PY.09.090171.001423
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Håkansson S, Galyov EE, Rosqvist R, Wolf-Watz H (1996) The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. 20:593–603. https://doi.org/10.1046/j.1365-2958.1996.5251051.x
Joardar V et al (2005) Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol 187:6488–6498
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJJN (2015) The Phyre2 web portal for protein modeling, prediction and analysis 10:845
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307. https://doi.org/10.5555/uri:pii:002221435490222X
Krueger, F (2012) Trim Galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Accessed 28/04/2016
Lindeberg M et al (2005) Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III hop effector proteins in the plant pathogen Pseudomonas syringae. Mol Plant–Microbe Interact 18:275–282. https://doi.org/10.1094/MPMI-18-0275
Mansfield J, Jenner C, Hockenhull R, Bennett MA, Stewart R (1994) Characterization of avrPphE, a gene for cultivar-specific avirulence from Pseudomonas syringae pv. phaseolicola which is physically linked to hrpY, a new hrp gene identified in the halo-blight bacterium. Mol Plant–Microbe Interact 7:726–739
Nimchuk ZL, Fisher EJ, Desveaux D, Chang JH, Dangl JL (2007) The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N-terminal domain for function. Mol Plant–Microbe Interact 20:346–357. https://doi.org/10.1094/MPMI-20-4-0346
Noble TJ, Tao Y, Mace ES, Williams B, Jordan DR, Douglas CA, Mundree SG (2017) Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Front Plant Sci 8:2102. https://doi.org/10.3389/fpls.2017.02102
Noble TJ, Young AJ, Douglas CA, Williams B, Mundree S (2019) Diagnosis and management of halo blight in Australian mungbeans: a review. Crop Pasture Sci 70:195–203. https://doi.org/10.1071/cp18541
Ryley M, Douglas C, Ryan M, Tatnell J, Martin W, King K, Keller L (2010) Integrated management of foliar pathogens of mungbean in Australia. In: Proceedings 1st Australian summer grains conference’. Gold Coast, Qld, pp 1–9
Shanmugasundaram S, Keatinge J, Hughes J (2010) The mungbean transformation: diversifying crops. Proven successes in Agricultural Development defeating malnutrition. Proven Successes in Agricultural Development, 381.
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EMJB (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. 31:3210–3212
Stevens C, Bennett MA, Athanassopoulos E, Tsiamis G, Taylor JD, Mansfield JW (1998) Sequence variations in alleles of the avirulence gene avrPphE.R2 from Pseudomonas syringae pv. phaseolicola lead to loss of recognition of the AvrPphE protein within bean cells and a gain in cultivar-specific virulence. Mol Microbiol 29:165–177. https://doi.org/10.1046/j.1365-2958.1998.00918.x
Sun S, Zhi Y, Zhu Z, Jin J, Duan C, Wu X, Xiaoming W (2017) An emerging disease caused by Pseudomonas syringae pv. phaseolicola threatens mung bean production in China. Plant Dis 101:95–102. https://doi.org/10.1094/PDIS-04-16-0448-RE
Sundin GW, Demezas DH, Bender CL (1994) Genetic and plasmid diversity within natural populations of Pseudomonas syringae with various exposures to copper and streptomycin bactericides. Appl Environ Microbiol 60:4421–4431
Taylor JD, Teverson DM, Allen DJ, Pastor-Corrales MA (1996a) Identification and origin of races of Pseudomonas syringae pv. phaseolicola from Africa and other bean growing areas. Plant Pathol 45:469–478. https://doi.org/10.1046/j.1365-3059.1996.d01-147.x
Taylor JD, Teverson DM, Allen DJ, Pastor Corrales MA (1996b) Identification and origin of races of Pseudomonas syringae pv. Phaseolicola from Africa and other bean growing areas. Plant Pathology (United Kingdom) 3 45(43):469–478. https://doi.org/10.1046/j.1365-3059.1996.d01-147.x
Tock AJ et al (2017) Genome-wide linkage and association mapping of halo blight resistance in common bean to race 6 of the globally important bacterial pathogen. Front Plant Sci 8:1170. https://doi.org/10.3389/fpls.2017.01170
Tsiamis G et al (2000) Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J 19:3204–3214. https://doi.org/10.1093/emboj/19.13.3204
Versalovic J, Koeuth T, Lupski R (1991) Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res 19:6823–6831. https://doi.org/10.1093/nar/19.24.6823
Versalovic J, Schneider M, Bruijn FJ, Lupski JR (1994) Genomic fingerprint of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5:25–40
Weingart H, Völksch B (1997) Genetic fingerprinting of Pseudomonas syringae pathovars using ERIC-, REP-, and IS50-PCR. J Phytopathol 145:339–345. https://doi.org/10.1111/j.1439-0434.1997.tb00411.x
Yoon S-H, Ha S-m, Lim J, Kwon S, JJAvL C (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. 110:1281–1286
Acknowledgments
This work was supported by the Australian Mungbean Association PhD scholarship program (2017000885) and the Centre for Tropical Crops and Biocommodities, Queensland University of Technology, Brisbane, Qld. The Grains and Research and Development Corporation (GRDC) and The Department of Agriculture and Fisheries of Queensland, Australia investments in grains pathology project (DAQ00186) and the National Mungbean Improvement Program (DAQ00210). We acknowledge the Office of eResearch at QUT for providing access to compute infrastructure and bioinformatics pipelines for this study. We also thank the industry as a whole for their support in providing samples from across Australia. (Burkholder 1926).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 206 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Noble, T.J., Young, A.J., Kelly, L.A. et al. Characterisation of the Pseudomonas savastanoi pv. phaseolicola population found in Eastern Australia associated with halo blight disease in Vigna radiata. Australasian Plant Pathol. 49, 515–524 (2020). https://doi.org/10.1007/s13313-020-00722-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-020-00722-8