Abstract
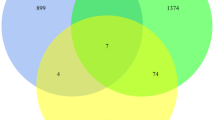
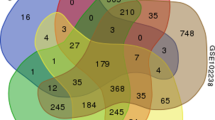
The study aimed to dissect the molecular mechanism of pancreatic cancer by a range of bioinformatics approaches. Three microarray datasets (GSE32676, GSE21654, and GSE14245) were downloaded from Gene Expression Omnibus database. Differentially expressed genes (DEGs) with logarithm of fold change (|logFC|) >0.585 and p value <0.05 were identified between pancreatic cancer samples and normal controls. Transcription factors (TFs) were selected from the DEGs based on TRASFAC database. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed for the DEGs using The Database for Annotation, Visualization and Integrated Discovery (p value <0.05), followed by construction of protein-protein interaction (PPI) network using Search Tool for the Retrieval of Interacting Genes software. Latent pathway identification analysis was applied to analyze the DEGs-related pathways crosstalk and the pathways with high weight value were included in the pathway crosstalk network using Cytoscape. Sixty-five DEGs were screened out. CCAAT/enhancer-binding protein delta (CEBPD), FBJ osteosarcoma oncogene B (FOSB), Stratifin (SFN), Krüppel-like factor 5 (KLF5), Pentraxin 3 (PTX3), and nuclear receptor subfamily 4, group A, member 3 (NR4A3) were important TFs. Interleukin-6 (IL-6) was the hub node of the PPI network. DEGs were significantly enriched in NOD-like receptor signaling pathway which was primarily interacted with inflammation and immune related pathways (cytosolic DNA-sensing, hematopoietic cell lineage, intestinal immune network for IgA production and chemokine pathways). The study suggested CEBPD, FOSB, SFN, KLF5, PTX3, NR4A3, IL-6, and NOD-like receptor pathways were involved in pancreatic cancer.



Similar content being viewed by others
Abbreviations
- TF:
-
transcription factor
- IL-6:
-
interleukin-6
- IL-1B:
-
interleukin-1β
- KRAS:
-
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
- TNF-α:
-
tumor necrosis factor-alpha
- EGF:
-
epidermal growth factor
- RMA:
-
robust multiarray average
- LogFC:
-
logarithm of fold change
- DAVID:
-
The Database for Annotation, Visualization, and Integrated Discovery
- GO:
-
gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- BP:
-
biological process
- CC:
-
cellular component
- MF:
-
molecular function
- STRING:
-
The Search Tool for the Retrieval of Interacting Genes
- PPI:
-
protein-protein interaction
- LPIA:
-
latent pathway identification analysis
- DEGs:
-
differentially expressed genes
- IFN-γ:
-
interferon-γ
- SFN:
-
stratifin
- FOSB:
-
FBJ osteosarcoma oncogene B
- PTX3:
-
pentraxin-related protein 3
- NR4A3:
-
nuclear receptor subfamily 4 group A, member 3
- C/EBP delta:
-
CCAAT/enhancer-binding protein delta
- KLFs:
-
Krüppel-like factors
- NLRs:
-
NOD-like receptors
- MAPK:
-
mitogen-activated protein kinase
References
Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137–50.
Seufferlein T, Bachet J, Van Cutsem E, Rougier P. Pancreatic adenocarcinoma: ESMO–ESDO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23 suppl 7:vii33–40.
Loos M, Kleeff J, Friess H, Büchler MW. Surgical treatment of pancreatic cancer. Ann N Y Acad Sci. 2008;1138(1):169–80.
Biankin AV, Waddell N, Kassahn KS, Gingras M-C, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405.
Maupin KA, Sinha A, Eugster E, Miller J, Ross J, Paulino V, et al. Glycogene expression alterations associated with pancreatic cancer epithelial-mesenchymal transition in complementary model systems. PLoS One. 2010;5(9):e13002.
Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH, et al. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology. 2010;138(3):949–57. e7.
Huang M, Tang S-N, Upadhyay G, Marsh JL, Jackman CP, Shankar S, et al. Embelin suppresses growth of human pancreatic cancer xenografts, and pancreatic cancer cells isolated from KrasG12D mice by inhibiting Akt and sonic hedgehog pathways. PLoS One. 2014;9(4):e92161.
Donahue TR, Tran LM, Hill R, Li Y, Kovochich A, Calvopina JH, et al. Integrative survival-based molecular profiling of human pancreatic cancer. Clin Cancer Res. 2012;18(5):1352–63.
Pang H, Lin A, Holford M, Enerson BE, Lu B, Lawton MP, et al. Pathway analysis using random forests classification and regression. Bioinformatics. 2006;22(16):2028–36.
Li Y, Agarwal P, Rajagopalan D. A global pathway crosstalk network. Bioinformatics. 2008;24(12):1442–7.
Basu A, Castle VP, Bouziane M, Bhalla K, Haldar S. Crosstalk between extrinsic and intrinsic cell death pathways in pancreatic cancer: synergistic action of estrogen metabolite and ligands of death receptor family. Cancer Res. 2006;66(8):4309–18.
Young SH, Rozengurt E. Crosstalk between insulin receptor and G protein-coupled receptor signaling systems leads to Ca (2) + oscillations in pancreatic cancer PANC-1 cells. Biochem Biophys Res Commun. 2010;401(1):154–8. doi:10.1016/j.bbrc.2010.09.036.
Maniati E, Bossard M, Cook N, Candido JB, Emami-Shahri N, Nedospasov SA, et al. Crosstalk between the canonical NF-κB and Notch signaling pathways inhibits Pparγ expression and promotes pancreatic cancer progression in mice. J Clin Invest. 2011;121(121):4685–99. 12.
Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80.
Leek JT. Surrogate variable analysis: University of Washington; 2007
Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9(4):326–32.
Dennis Jr G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):3.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9.
Arakawa K, Kono N, Yamada Y, Mori H, Tomita M. KEGG-based pathway visualization tool for complex omics data. In silico biol. 2005;5(4):419–23.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(D1):D808–D15.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Pham L, Christadore L, Schaus S, Kolaczyk ED. Network-based prediction for sources of transcriptional dysregulation using latent pathway identification analysis. Proc Natl Acad Sci. 2011;108(32):13347–52.
Becker AE, Hernandez YG, Frucht H, Lucas AL. Pancreatic ductal adenocarcinoma: risk factors, screening, and early detection. World J Gastroenterol : WJG. 2014;20(32):11182–98. doi:10.3748/wjg.v20.i32.11182.
Moore F, Santin I, Nogueira TC, Gurzov EN, Marselli L, Marchetti P, et al. The transcription factor C/EBP delta has anti-apoptotic and anti-inflammatory roles in pancreatic beta cells. PLoS One. 2012;7(2):e31062.
Kim JH, Lee JY, Lee KT, Lee JK, Lee KH, Jang K-T, et al. RGS16 and FosB underexpressed in pancreatic cancer with lymph node metastasis promote tumor progression. Tumor Biol. 2010;31(5):541–8.
Guweidhi A, Kleeff J, Giese N, El Fitori J, Ketterer K, Giese T, et al. Enhanced expression of 14-3-3sigma in pancreatic cancer and its role in cell cycle regulation and apoptosis. Carcinogenesis. 2004;25(9):1575–85.
Mori A, Moser C, Lang SA, Hackl C, Gottfried E, Kreutz M, et al. Up-regulation of Kruppel-like factor 5 in pancreatic cancer is promoted by interleukin-1beta signaling and hypoxia-inducible factor-1alpha. Mol Cancer Res : MCR. 2009;7(8):1390–8. doi:10.1158/1541-7786.mcr-08-0525.
Locatelli M, Ferrero S, Martinelli Boneschi F, Boiocchi L, Zavanone M, Maria Gaini S, et al. The long pentraxin PTX3 as a correlate of cancer-related inflammation and prognosis of malignancy in gliomas. J Neuroimmunol. 2013;260(1):99–106.
Lee S-O, Abdelrahim M, Yoon K, Chintharlapalli S, Papineni S, Kim K, et al. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Res. 2010;70(17):6824–36.
Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11(3):291–302.
Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1):349–61.
Dima SO, Tanase C, Albulescu R, Herlea V, Chivu-Economescu M, Purnichescu-Purtan R, et al. An exploratory study of inflammatory cytokines as prognostic biomarkers in patients with ductal pancreatic adenocarcinoma. Pancreas. 2012;41(7):1001–7.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Rincon M. Interleukin-6: from an inflammatory marker to a target for inflammatory diseases. Trends Immunol. 2012;33(11):571–7.
Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19(4):456–69.
Guan J, Zhang H, Wen Z, Gu Y, Cheng Y, Sun Y et al. Retinoic acid inhibits pancreatic cancer cell migration and EMT through the downregulation of IL-6 in cancer associated fibroblast cells. Cancer letters. 2013.
Hong DS, Angelo LS, Kurzrock R. Interleukin‐6 and its receptor in cancer. Cancer. 2007;110(9):1911–28.
Song J, Singh M. How and when should interactome-derived clusters be used to predict functional modules and protein function? Bioinformatics. 2009;25(23):3143–50.
Fukata M, Vamadevan AS, Abreu MT, editors. Toll-like receptors (TLRs) and Nod-like receptors (NLRs) in inflammatory disorders. Seminars in immunology; 2009: Elsevier.
Huzarski T, Lener M, Domagała W, Gronwald J, Byrski T, Kurzawski G, et al. The 3020insC allele of NOD2 predisposes to early-onset breast cancer. Breast Cancer Res Treat. 2005;89(1):91–3.
Papaconstantinou I, Theodoropoulos G, Gazouli M, Panoussopoulos D, Mantzaris GJ, Felekouras E, et al. Association between mutations in the CARD15/NOD2 gene and colorectal cancer in a Greek population. Int J Cancer. 2005;114(3):433–5.
Tsuji M, Suzuki K, Kinoshita K, Fagarasan S, editors. Dynamic interactions between bacteria and immune cells leading to intestinal IgA synthesis. Seminars in immunology; 2008: Elsevier.
Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27(2):373–83. doi:10.1038/sj.emboj.7601962.
Park J-H, Kim Y-G, McDonald C, Kanneganti T-D, Hasegawa M, Body-Malapel M, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178(4):2380–6.
Acknowledgments
This study was supported by Overseas Scholars Funds (grant no. 1054HQ081) and National Natural Science Foundation of China (grant no. 30340058).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Highlights
1. We found 65 DEGs from gene expression profiles of 78 pancreatic cancer specimens.
2. This study identified 6 significant TFs (CEBPD, FOSB, SFN, KLF5, PTX3, and NR4A3).
3. NOD-like receptor pathway was suggested to play a key role in pancreatic cancer.
4. Pathway crosstalk between NOD-like receptor pathway and multiple inflammation and immune related pathways was predicted.
The Publisher and Editor retract this article in accordance with the recommendations of the Committee on Publication Ethics (COPE). After a thorough investigation we have strong reason to believe that the peer review process was compromised.
An erratum to this article is available at http://dx.doi.org/10.1007/s13277-015-3788-1.
About this article
Cite this article
Liu, J., Li, J., Li, H. et al. RETRACTED ARTICLE: A comprehensive analysis of candidate genes and pathways in pancreatic cancer. Tumor Biol. 36, 1849–1857 (2015). https://doi.org/10.1007/s13277-014-2787-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2787-y