Abstract
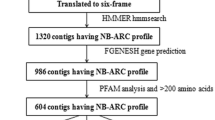
As the largest class of resistant genes, the nucleotide binding site (NBS) has been studied extensively at a genome-wide level in rice, sorghum, maize, barley and hexaploid wheat. However, no such comprehensive analysis has been conducted of the NBS gene family in Triticum urartu, the donor of the A genome to the common wheat. Using a bioinformatics method, 463 NBS genes were isolated from the whole genome of T. urartu, of which 461 had location information. The expansion pattern and evolution of the 461 NBS candidate proteins were analyzed, and 118 of them were duplicated. By calculating the lengths of the copies, it was inferred that the NBS resistance gene family of T. urartu has experienced at least two duplication events. Expression analysis based on RNA-seq data found that 6 genes were differentially expressed among Tu38, Tu138 and Tu158 in response to Blumeria graminis f.sp.tritici (Bgt). Following Bgt infection, the expression levels of these genes were up-regulated. These results provide critical references for further identification and analysis of NBS family genes with important functions.




Similar content being viewed by others
References
Akhunov ED, Sehgal S, Liang H, Wang S, Akhunova AR, Kaur G, Li W, Forrest KL, See D, Simková H et al (2013) Comparative analysis of syntenic genes in grass genomesreveals accelerated rates of gene structure and coding sequence evolutionin polyploid wheat. Plant Physiol 161:252–265
Arya P, Kumar G, Acharya V, Singh AK (2014) Genome-wide identification and expression analysis of NBS-encoding genes in Malus x domestica and expansion of NBS genes family in Rosaceae. PLoS ONE 9:e107987
Aswati Nair R, Thomas G (2007) Isolation, characterization and expression studies of resistance gene candidates (RGCs) from Zingiber spp. Theor Appl Genet 116:123–134
Azhar M, Heslop-Harrison JS (2008) Genomes, diversity and resistance gene analogues in Musa species. Cytogenet Genome Res 121:59–66
Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE et al (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12:1871–1884
Botella MA, Parker JE, Frost LN, Bittner-Eddy PD, Beynon JL, Daniels MJ, Holub EB, Jones JD (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10:1847–1860
Bouktila D, Khalfallah Y, Habachi-Houimli Y, Mezghani-Khemakhem M, Makni M, Makni H (2014) Large-scale analysis of NBS domain-encoding resistance gene analogs in Triticeae. Genet Mol Biol 37:598–610
Burland TG (2000) DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol 132:71–91
Cannon SB, Zhu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54:548–562
Cheng Y, Li XY, Jiang HY, Ma W, Miao WY, Yamada T, Zhang M (2012) Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J 279:2431–2443
Ellis JG, Lagudah ES, Spielmeyer W, Dodds PN (2014) The past, present and future of breeding rust resistant wheat. Front Plant Sci 5:641
Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial resistance gene is a member of the TIR-NBS-LRR family of disease resistance genes. Plant J 20:265–277
Gaut BS, Conery JF (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94:6809–6814
Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH (2002) Extent of gene duplication in the genomes of Drosophila, nematode and yeast. Mol Biol Evol 19:256–262
Gu LJ, Si WN, Zhao LN, Yang S, Zhang X (2015) Dynamic evolution of NBS-LRR genes in bread wheat and its progenitors. Mol Genet Genomic 290:727–738
Guo Y, Qiu LJ (2013) Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 8: e76809
Jia RZ, Ming R, Zhu YJ (2013) Genome-Wide analysis of nucleotide-binding site (NBS) disease resistance (R) genes in Sacred Lotus (Nelumbo nucifera Gaertn.) reveals their transition role during early evolution of land plants. Trop Plant Biol 6:98–116
Jia YX, Yuan Y, Zhang YC, Yang SH, Zhang XH (2015) Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet 16:48
Li J, Ding J, Zhang W, Zhang YL, Tang P, Chen JQ, Tian DC, Yang SH (2010) Unique evolutionary pattern of numbers of gramineous NBS–LRR genes. Mol Genet Genomics 283:427–438
Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y et al (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496:87–90
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Ma H, SinghA RP, Mujeeb-Kazi A (1997) Resistance to stripe rust in durum wheats, A-genome diploids, and their amphiploids. Euphytica 94:279–286
Mackey D, Holt BF, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108:743–754
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Parker JE, Coleman MJ, Szabò V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, Jones JD (1997) The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 9:879–894
Pollier J, Rombauts S, Goossens A (2013) Analysis of RNA-Seq data with TopHat and Cufflinks for genome-wide expression analysis of jasmonate-treated plants and plant cultures. Methods Mol Biol 1011:305–315
Porter BW, Paidi M, Ming R, Alam M, Nishijima WT, Zhu YJ (2009) Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Gen Genomics 281:609–626
Qiu YC, Zhou RH, Kong XY, Zhang SS, Jia JZ (2005) Microsatellite mapping of a Triticum urartu Tum. derived powdery mildew resistance gene transferred to common wheat (Triticum aestivum L.). Theor Appl Genet 111:1524–1531
Repet WL, Feldman GD (1991) The earliest remains of grasses in the fossil record. Am J Bot 78:1010–1014
Richter TE, Ronald PC (2000) The evolution of disease resistance genes. Plant Mol Biol 42:195–204
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Rouse MN, Jin Y (2011) Stem rust resistance in A-genome diploid relatives of wheat. Plant Dis 95:941–944
Sanseverino W, Hermoso A, D’Alessandro R, Vlasova A, Andolfo G, Frusciante L, Lowy E, Roma G, Ercolano MR (2012) PRGdb 2.0: towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res 41:1167–1171
Srichumpa P, Brunner S, Keller B, Yahiaoui N (2005) Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 139:885–895
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:609–612
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tarr DE, Alexander HM (2009) TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res Notes 2:197
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tommasini L, Yahiaoui N, Srichumpa P, Keller B (2006) Development of functional markers specific for seven Pm3 resistance alleles and their validation in the bread wheat gene pool. Theor Appl Genet 114:165–175
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-SEq. Bioinformatics 25:1105–1111
Wang XY, Chen J, Yang Y, Zhou J, Qiu Y, Yu CL, Cheng Y, Yan CH, Chen JP (2013) Characterization of a novel NBS-LRR gene involved in bacterial blight resistance in rice. Plant Mol Biol Rep 31:649–656
Wei HL, Li W, Sun XW, Zhu SJ, Zhu J (2013) Systematic analysis and comparison of nucleotide-binding site disease resistance genes in a diploid cotton Gossypium raimondii. PLoS ONE 8:e68435
Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78:1101–1115
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37:528–538
Zhang H, Yang YZ, Wang CY, Liu M, Li H, Fu Y, Wang YJ, Nie YB, Liu XL, Ji WQ (2014) Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics 15:898
Zhao Y, Weng QY, Song JH, Ma HL, Yuan JC, Dong ZP, Liu YH (2016) Bioinformatics analysis of NBS-LRR encoding resistance genes in Setaria italica. Biochem Genet 54:232–248
Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Gen. Genomics 271:402–415
Acknowledgements
This project is funded by Shanxi Province Science Foundation for Youths (2015021145), Shanxi Province Finance-supported Agricultural Project (2015ZYZX-03), Shanxi Province Technologies R & D Progaram (20150311001-1), Shanxi Province International Cooperation Project (201603D421003), and Technologies R&D Program of the Shanxi Academy of Agricultural Sciences (YGG1602, 15YGG01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Jing Liu, Linyi Qiao, Xiaojun Zhang, Xin Li, Haixian Zhan, Huijuan Guo, JunZheng and Zhijian Chang declare no conflict of interest on the contents in the manuscript.
Ethical approval
This article does not contain any studies with human subjects or animals performed by any of the authors.
Additional information
Jing Liu and Linyi Qiao have equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, J., Qiao, L., Zhang, X. et al. Genome-wide identification and resistance expression analysis of the NBS gene family in Triticum urartu . Genes Genom 39, 611–621 (2017). https://doi.org/10.1007/s13258-017-0526-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-017-0526-7