Abstract
COVID-19 is an infectious disease, which has adversely affected public health and the economy across the world. On account of the highly infectious nature of the disease, rapid automated diagnosis of COVID-19 is urgently needed. A few recent findings suggest that chest X-rays and CT scans can be used by machine learning for the diagnosis of COVID-19. Herein, we employed semi-supervised learning (SSL) approaches to detect COVID-19 cases accurately by analyzing digital chest X-rays and CT scans. On a relatively small COVID-19 radiography dataset, which contains only 219 COVID-19 positive images, 1341 normal and 1345 viral pneumonia images, our algorithm, COVIDCon, which takes advantage of data augmentation, consistency regularization, and multicontrastive learning, attains 97.07% average class prediction accuracy, with 1000 labeled images, which is 7.65% better than the next best SSL method, virtual adversarial training. COVIDCon performs even better on a larger COVID-19 CT Scan dataset that contains 82,767 images. It achieved an excellent accuracy of 99.13%, at 20,000 labels, which is 6.45% better than the next best pseudo-labeling approach. COVIDCon outperforms other state-of-the-art algorithms at every label that we have investigated. These results demonstrate COVIDCon as the benchmark SSL algorithm for potential diagnosis of COVID-19 from chest X-rays and CT-Scans. Furthermore, COVIDCon performs exceptionally well in identifying COVID-19 positive cases from a completely unseen repository with a confirmed COVID-19 case history. COVIDCon, may provide a fast, accurate, and reliable method for screening COVID-19 patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Novel coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).Footnote 1 Due to the highly infectious nature of the disease, it has spread worldwide in a short period of time, adversely affecting public health and global economy. In order to reduce the rapid spread of COVID-19, fast and accurate diagnosis of patients is of primary interest.
The definitive test for COVID-19 is the real-time reverse transcriptase polymerase chain reaction (RT-PCR) test, which has a sensitivity between 51 and 94% and takes around 24 hours to obtain the results [29].Footnote 2 The long waiting time may increase the chances of spreading the disease to others. CT scans and standard chest X-rays—on the other hand, may save time for the diagnosis of COVID-19 [74]. Cases have been reported where RT-PCR gave false-negative results, but radiographic techniques could detect COVID-19 [26, 67]. Chest X-rays and chest CT scans are, therefore, being used to get more information and are being considered a screening tool alongside the RT-PCR test [7, 33]. In fact, a study conducted by Ai et al. [3] shows that CT Scan has better sensitivity compared to RT-PCR in detecting COVID-19 and can be used for the diagnosis of the disease. CT-Scans can also provide information about the severity of COVID-19, which cannot be obtained by standard RT-PCR testing.
Chest CT scans and X-rays of COVID-19 patients often display abnormal patterns, for example, bilateral, multilobar ground glass opacities (GGO) with a peripheral or posterior distribution, mainly in the lower lobes and on occasion in the middle lobe [70]. The visual differences in chest CT scan and X-ray images between COVID-19, pneumonia, and normal patients are subtle and require expert radiologists, which causes a bottleneck as their number is limited. In this context, artificial intelligence/machine learning can help to identify positive cases, find abnormalities and provide aid to medical personnel.
For the past few years, deep learning models have demonstrated their potentials to be useful to radiologists and medical imaging experts for various disease detection and classification tasks such as skin cancer [16], brain disease [34], arrhythmia [52], breast cancer [39], pneumonia [48], etc. A few state-of-the-art supervised models have even achieved performance equivalent to experienced radiologists [48]. Recently, deep learning-based approaches have also been applied successfully in diagnosis of COVID-19 [22] . The extraordinary performance of such deep learning models based on supervised learning, however, requires large amounts of labeled data [11]. A situation that may be challenging in medical image analysis, where data collection and annotation are time-intensive tasks and increase the workload on the radiologists.
Semi-supervised learning (SSL) curbs the disadvantages of supervised learning methods by utilizing a small amount of labeled and a larger amount of unlabeled data [73]. Data augmentation, together with transfer learning, can produce powerful, more robust models that require less training time. In medical imaging, large unlabeled datasets are readily available along with smaller high-quality labeled datasets. Therefore, SSL methods can be an excellent option for automated medical image diagnosis.
Up until now, many SSL models, such as Pseudo-labelling, [32] VAT, [40] \(\pi\)-Model, [31] Mean Teacher, [55] MixMatch, [10] and FixMatch [53] have been implemented successfully for analyzing two-dimensional image data. Recently, we also reported a semi-supervised image classification algorithm, MultiCon [49], which uses multi-contrastive learning alongside consistency regularization to outperform other semi-supervised approaches in image classification.
Consistency regularization is a semi-supervised learning component that adds a supplementary loss function to a network such that the predictions of the network remain consistent even after the inputs are perturbed. This is done by making use of the unlabeled data to find latent features for the additional loss function. Phillip et al. [8] developed the first variant of this component which was later made popular by Samuli et al. [31] and Mehdi et al. [50] . Some of the recent variations include replacing parts of the loss function [40], using this component in larger SSL pipeline [10, 30] and replacing \(\ell ^2\) loss with cross-entropy [66].
Contrastive Learning is the process that allows models to learn high level features about the dataset by finding how similar or different a pair of data points are from each other. This is an unsupervised learning technique that is implemented before any segmentation or classification task. Despite being a relatively new area of study, it has been the basis for many excellent works such as contrastive predictive coding [20, 43], representation learning using Deep InfoMax (DIM) [4, 21, 59] or momentum contast [18], learning invariances using Augmented Multiscale DIM [9] or Contrastive MultiView Coding [56] etc.
Previous deep learning approaches have shown promises in identifying COVID-19 cases from chest radiography images [5, 13, 14, 19, 22, 46, 61,62,63,64, 71, 72], please refer to Table 1. But as the performance of these methods depend on large labeled dataset, researchers have also tried to explore the performance of semi-supervised learning methods in the diagnosis of COVID-19 cases [2, 17, 27, 28, 36]. Jun et al. [36] used active contour regularization on a region scalable fitting (RSF) model to further tune the pseudo-labels of the unlabeled CT images. Mohammed et al. [2] proposed a dual path few shot semi-supervised segmentation approach that uses only a few labeled CT images to accurately segment COVID-19 infection. A semi-supervised shallow framework was proposed by Debanjan et al. [28] that diagnosed segmentation of CT images produced by parallel quantum-inspired self-supervised Network (PQIS-Net). Shahin et al. [27] used autoencoder based semi supervised approach to first extract regions of interest from chest X-ray images which are then fed to a deep architecture to classify them.
Herein, we use the state-of-the-art SSL algorithms and our algorithm, MultiCon [49], for the classification of grey-scale X-ray and CT scan images as COVID-19, pneumonia, or normal cases from a publicly available COVID-19 radiography dataset [47] and a COVID-19 CT Scan dataset [71]. In the present work, we refer MultiCon as COVIDCon for better understanding to the readers. The loss function of COVIDCon is constituted using two loss terms—consistency regularization and multi-contrastive learning Fig. 1. The consistency regularization component keeps the probability distributions of weakly augmented and strongly augmented dataset similar to each other. The multi-contrastive learning part keeps the data points of the same class together and instances of different classes further apart. The notion behind using these two components together is the complementary nature of these components. If consistency regularization is removed, the loss function would be optimized regardless of the labeled data. Similarly, if multi-contrastive learning is removed, the predictions would be inaccurate due to the scarce amount of labeled data.
The framework of COVIDCon. Different input chest X-rays with corresponding (weak and strong) augmented transformations are projected into embedding features. For the consistency regularization, the model prediction works as pseudo-label, \(\ell _{CR}\) aims to make the output from strong augmentation match of the pseudo-label. The model is trained under the combination of \(\ell _{CR}\) and \(\ell _{MC}\). The colored solid lined circles represent strong augmented data points, the dotted circles represent weak augmented data points, and same color represents same class. Dashed line is the probability distribution of weak augmented unlabeled images which are used for pseudo-labeling
Methodology
This section presents the methods and materials used in this study. Sections 2.1 and 2.2 refer to the descriptions of COVID-19 Radiography and CT Scan datasets used for training and testing the SSL methods.
COVID-19 radiography dataset
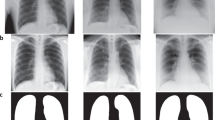
The COVID-19 radiography dataset [47] has been developed by a team of researchers from Qatar University, the University of Dhaka and their collaborators from Pakistan and Malaysia with the help of medical doctors. The dataset consists mostly of posterior-to-anterior images of chest X-rays with 1024 × 1024 resolution from COVID-19 positive cases, viral pneumonia cases, as well as normal cases. COVID-19 Radiography dataset is available publicly, and it gets updated continuously with new X-ray images. As of 14th June 2020, the dataset contains a total of 2905 unique images, which are distributed over 3 imbalanced classes, namely COVID-19, Normal and Viral Pneumonia. The COVID-19 class contains 219 unique X-ray images, while viral pneumonia and normal classes contain 1341, 1345 images, respectively. A few sample chest X-rays of the COVID-19, viral pneumonia, and normal cases are listed in Fig. 2.
COVID-19 CT scan dataset
The China Consortium of Chest CT Image Investigation (CC-CCII) compiled a dataset [71] of CT images, with 512 × 512 resolution, from cohorts of their patients. The dataset consists of 617,775 images, which are 2D slices, from 4154 patients divided into three classes of novel coronavirus pneumonia (NCP), common pneumonia (CP), and normal cases. The images in the common pneumonia class are a mixture of some of the common classes of pneumonia in China, such as viral pneumonia and bacterial pneumonia. The version used for this experiment (version 2.2) is publicly available for download and was released on 24th April 2020. Image slices containing lesions were only used to train the model, resulting in 21,777 images in the coronavirus pneumonia class and 36,894 images in the common pneumonia class. In order to maintain a balance among the data in the three classes, a subset of 24,096 images from the normal class was chosen randomly. We used 617,775 CT images from 4154 patients. The metadata.csv file (reference 54) contains the information that each patient is classified to individual categories such as: pneumonia (NCP), covid (CP) and normal. We took the advantage of this csv file and obtained the patient ids classified into different categories. From those patient ids we randomly chose 80% for our train data and rest 20% patient ids and corresponding scan ids in order to segregate between the train and test data. To avoid overfitting, we ensured that none of the patient gets assigned to both train and test data. A few sample CT scans of the COVID-19, common pneumonia and normal cases are listed in Fig. 3.
COVID-19 unseen dataset
The COVID-19 unseen datasetFootnote 3 has been assembled and maintained by the The American College of Radiology (ACR). The dataset contains 51 images in total each of which are from unique patients diagnosed with COVID-19.
Our algorithm: COVIDCon
For the detection of COVID-19 from the digital chest X-Rays and CT scans, we have used our algorithm, COVIDCon, which is based on our previously developed algorithm [49]. We have compared the performance of COVIDCon with other state-of-the-art SSL methods including MixMatch [10], Virtual Adversarial Training (VAT) [40], Pseudo-labeling [32], Mean Teacher [55], \(\varPi\) model [31], Interpolation Consistency Training (ICT) [60] in (Table 2).
Though we applied MultiCon [49] in the field of drug classification, the use of contrastive learning is still relatively unexplored in the field of medical imaging. We observed that, similar to drug classification, X-ray and CT image classification, especially in the case of COVID-19, is mostly dependent on discerning subtle features. MultiCon’s impressive performance in drug classification inspired us to apply it to the medical imaging domain as COVIDCon.
COVIDCon is a semi-supervised learning (SSL) technique that combines consistency regularization and multi-contrastive learning approach to learn a feature embedding, where the augmented views of the same data maintain a minimum distance between them. The main inspiration behind such a technique is to allow deep neural network models to train with the minimal number of labeled data and a large unlabeled dataset. This is especially advantageous since unlabeled data is cheap and easy to obtain, and it also relieves experts from manually labeling them. The proposed algorithm is a three-step process which comprises of augmenting the data, followed by pseudo-labelling the unlabeled data and then finally obtaining a feature representation using a contrastive prediction task. Unlike other methods, COVIDCon combines the loss terms from both consistency regularization and multi-contrastive learning so that the feature embedding can satisfy the positive concentrated and negative separated properties.
Data augmentations used in this technique are of two types: weak augmentation (\(\mathcal {W}_{a}\)) and strong augmentation \(\mathcal {S}_{a}\). Weak augmentation corresponds to a flip-shift strategy, which randomly flips images with a probability of 50% and translates them vertically and horizontally up to 12.5%. Strong augmentation strategies are based on RandAugment [53]. We use color inversion, contrast adjustment, translation, transformations. RandAugment selects a subset of these and randomly assigns how intense they will be within a mini-batch.
A side effect of using strong augmentations, is that the image might differ a lot from the original image. For this reason, the weakly augmented version of each unlabeled image is assigned a pseudo label and the model is optimized using cross entropy loss by assigning the above pseudo label to the corresponding strongly augmented version. The supervised cross entropy loss of labeled images and the unsupervised loss mentioned above combine together to make consistency regularization. Consistency regularization ensures the fact that even after the perturbations caused by strong augmentation, the labeled and unlabeled data follow a similar distribution. Contrastive learning maximizes the mutual information between the differently augmented views of the data. The idea behind contrastive learning is to learn representations such that similar samples stay close to each other, while dissimilar ones are far apart. This results in separation of classes where samples form clusters on the basis of their classes in the feature space, which consequently results in improved predictions.
After assigning a pseudo label to strongly augmented unlabeled images, we select those images in the mini-batch whose pseudo labels are predicted with a probability greater than a threshold. Of the selected images, we extract their feature vectors and reduce distances between vectors of images with same class while increasing distances between vectors of images with different classes. This strategy results in clustering of images with same classes and results in a more generalized model.
where \(q_u\) are the probabilities of the labels of the unlabeled image, \(max(q_u)\) is the probability of the pseudo label, \(\tau\) is the threshold, \(y_l\) is the label for the labeled image, \(y_u\) is the correct label of the unlabeled image, \(\hat{y}_u\) is the pseudo label of the unlabeled image, \(\hat{y}_l\) is the actual label of the labeled image.
In the case of multi-contrastive learning, the augmented dataset is clustered with the goal of grouping data from the same class and pushing the data from different classes further away from each other. Given the similarity measurement \(\mathcal {S}(x_{i},x_{j}) = \mathcal {S}(f(x_i;\theta ), f(x_j;\theta ))\) of the sample pair \((x_i, x_j)\), the multi-contrastive loss \(\ell _{MC}\) is:
where \(\omega\) is the hyperparameter in the Binomial deviance [69]. \(m^{+}\) is the number of positive pairs as designated by \(y_{i}=y_{j}\) and \(m^{-}\) is the number of negative pairs as designated by \(y_{i} \ne y_{j}\). Finally, the consistency regularization component and multi-contrastive loss sums together and gives the overall loss term \(\ell\): \(\ell = \ell _{CR} + \lambda \ell _{MC}\) Where \(\lambda\) is a fixed scalar hyperparameter denoting the relative weight of different objective function.
COVIDCon is a specialized algorithm developed for two-dimensional image analysis. It is especially suitable for identifying subtle visual differences in images and therefore has the potential in clinical diagnosis, for example, detection of COVID-19 by X-ray image analysis.
Implementation details
We utilized Pytorch Footnote 4 to implement COVIDCon on the radiography and CT Scan datasets. The X-ray and CT Scan images were resized into 84 × 84 and pretrained ResNeXt-101 [65] was used as the deep learning model. The ResNeXt model was pretrained in a weakly-supervised fashion on 940 million public images with 1.5K labels matching with 1000 ImageNet1K synsets, followed by fine-tuning on ImageNet1K dataset Footnote 5. For the implementation of COVIDCon, we randomly initialized and added 3 fully connected layers to ResNeXt. COVIDCon was trained for 10 epochs, the learning rate was set at 0.01, the size of the mini-batch was 128, and the weight decay was 0.001.
We used an identical set of hyperparameters (\(\lambda _{1}\) = 2, \(\lambda _{2}\) = 40, \(\omega\) = 1 and \(\eta\) = 0.1, \(\lambda = 0.8\) during the training step) for all experiments. These hyperparameters are chosen in such a way that they empirically gave convergent results. For training and testing, we split the dataset into 80:20 ratio, respectively. To avoid any overfitting, we ensured that none of the image gets assigned to both train and test data. Overall, we repeat each experiment five times independently and report the average result.
Baseline methods
We consider recent state-of-the-art methods, such as \(\varPi\)-Model [31], Mean Teacher [55], ICT [60], Virtual Adversarial Training [40], Pseudo-Label [32], MixMatch [10], and FixMatch, [53] as the baseline methods. We use the official codes from the original papers and implement the same network architecture ResNeXt-101, [65] training epoch, and initialized the hyperparameters in each method based on the author’s recommendations. We retuned the hyperparameters for each baseline method to ensure a fair comparison with COVIDCon.
Results
Results on COVID-19 radiography dataset
We first employed COVIDCon on the COVID-19 Radiography dataset and obtained the average class prediction accuracies (Table 3) of the model with different numbers of labeled data. COVIDCon outperformed other state-of-the-art SSL methods at all labeled data. With just 50 labels, COVIDCon achieved an average accuracy of 92.71%, which is slightly better than the next best state-of-the-art FixMatch model. The performance of FixMatch, however, decreased with more labeled data on account of overfitting, an issue, which was not observed in the case of COVIDCon. COVIDCon reached the highest class prediction accuracy of 97.07% on average with 1000 labeled data, which is 7.65% better than the next best VAT method.
In order to better understand the performance of COVIDCon, we analyzed (Fig. 4) the confusion matrix. For 98.73% of cases, COVIDCon correctly predicted the true class for COVID-19 X-ray images with 1000 labeled data. The prediction accuracies were 96.1% and 97.6% for normal and viral pneumonia cases, respectively. At 1000 labels, FixMatch, the next best state-of-the-art, obtained accuracies on identifying COVID-19 and viral pneumonia cases with 82.60% and 91.9%, respectively. Almost 9 COVID cases have been misclassified as viral pneumonia at these labels. Given the small and imbalanced nature of the dataset, COVIDCon performed well in predicting all the three classes. It successfully differentiated all COVID-19 cases from viral pneumonia cases and confused only 1.27% COVID-19 cases as normal. Given the similarities of the image features in some of the chest X-rays of COVID-19 and viral pneumonia cases, COVIDCon performed well, and a few mispredictions are well within the limit of diagnostic inaccuracy in actual clinical settings.
Confusion matrices from a COVIDCon and b FixMatch showing the proportion of each predicted class (x-axis) for chest X-ray images in each true class (y-axis) with 1000 labels on the COVID-19 Radiography dataset. True class prediction accuracies are highlighted in bold. All numbers are rounded to two decimal places
The class prediction accuracies are improved with increasing epoch numbers. At 6 epoch COVIDCon reached the accuracy of 94.22%, which is slightly decreased at 10 epoch with 50 labeled data.
Results on COVID-19 CT scan dataset
Confusion matrices from a COVIDCon and b VAT showing the proportion of each predicted class (x-axis) for chest CT scan images in each true class (y-axis) with 5000, 20,000 labels on the COVID-19 CT Scan dataset, and true class prediction accuracies are highlighted in bold. All numbers are rounded to two decimal places
The class prediction accuracies are improved with increasing epoch numbers. At 6 epoch COVIDCon reached the accuracy of 98.56%, which increased further to 99.13% at 10 epoch with 20,000 labeled data. The confusion matrices in Fig. 5 show that COVIDCon achieved excellent accuracies to identify all of the three classes. With increasing labeled data, independent class prediction accuracies have also increased. For example, with 5000 labeled data, COVIDCon misclassified 3 out of 100 COVID-19 cases as viral pneumonia, whereas just 1.6% COVID-19 cases were classified wrongly as viral pneumonia. At 5000 and 20000 labels, VAT, the next best state-of-the-art, misclassified almost 8 and 11 COVID cases as viral pneumonia, respectively. Therefore, COVIDCon is more accurate compared to other state of the art.
Receiver operator characteristics curve
Receiver operator characteristic areas under the curves (ROC-AUC) are obtained (Fig. 6) on the COVID-19 Radiography and COVID-19 CT scan datasets at 1000 and 20,000 labels, respectively. The microaverage Area Under the Curve (AUC) is found to be 0.99 and 1, respectively, for both the datasets. The ROC-AUC values for individual classes lie in the range of 0.98 and 1, proving the efficacy of our method.
Consistent with the results obtained on the radiography dataset, COVIDCon outperformed other state-of-the-art SSL methods at all numbers of labeled data. The performance of COVIDCon on the CT scan dataset is recorded in Table 4. With 5000 labeled data, COVIDCon achieved an average accuracy of 98.30%, which is 6% better than the next best state-of-the-art Pseudo Label model. COVIDCon reached the highest-class prediction accuracy of 99.13% on average, with 20,000 labeled data, which is 6.45% better than the next best state of the art model, Mix Match.
t-Distributed stochastic neighbor embedding (t-SNE) analysis
In order to understand the discrimination of learned embedding from our approach, we visualize the final embedding using t-SNE [37] implementation, which shows a snapshot of the COVID-19 Radiography dataset projected into a 2-dimensional feature space. We mimic the learning procedure by randomly selecting 1000 labeled samples under 10 epochs training. Then we test the learned embedding on the test set and show (Fig. 7) the visualizations. The results demonstrate the fact that with the assistance of multi-contrastive loss, COVIDCon could reduce the intra-class variances much better in comparison to the next best FixMatch approach, and produce well-separated feature embeddings, resulting in high prediction accuracy for all of the three classes as can be seen from the confusion matrices in Figs. 4 and 5. Similarly, COVIDCon produces much better well-separated feature embeddings (Fig. 7) on CT scan datasets as well. All these results demonstrate the importance of the multi-contrastive loss component (Table 2) of COVIDCon.
Testing COVIDCon on COVID-19 unseen dataset
Oftentimes machine learning model that works almost perfectly using cross-validation fails miserably when tested on new unseen data. Therefore we tested COVIDCon on a small repositoryFootnote 6 of confirmed COVID-19 cases, where each data instance has a case history. COVIDCon performed well in identifying COVID-19 positive cases with 98% accuracy. It correctly identified all cases in the dataset except for the case number 59,638 Footnote 7 (Fig. 8, which is predicted as normal from chest X-ray analysis instead of positive. Furthermore, cases numbered 59,554 and 56,442 having unknown COVID-19 test result, are also identified as positive by COVIDCon. Interestingly, the case 56442 has COVID-suspected CT features (Fig. 8), and is also identified successfully as COVID-19 positive by COVIDCon. These results demonstrate the accuracy of our model and its potential use in the clinical settings.
Ablation study
In order to understand the effect of each loss component of COVIDCon, we performed an ablation study. We isolated different components of our loss function and investigated their impacts on the model’s performance. We evaluated supervised cross-entropy (CE), consistency regularization (CR), cross-entropy and multi-contrastive (MC) loss, and finally COVIDCon. From the prediction accuracies for the COVID-19 Radiography and CT Scan datasets, as recorded in Table 5, we observed that the contrastive loss in conjunction with supervised cross-entropy performed quite well on its own. But the composition of all components, proposed in COVIDCon demonstrated the best performance.
Visualization of learned feature embeddings on COVID-19 Radiography and CT Scan datasets. a COVIDCon, b FixMatch feature embedding with 1000 labels on COVID-19 Radiography dataset. c COVIDCon, d FixMatch feature embedding with 20,000 labels on CT Scan dataset. Red, blue, and green dots are COVID-19, pneumonia, and normal cases
X-rays and CT scan cases from a new unseen COVID-19 Repository (https://cortex.acr.org/COVID19/). a Case number 59,638: chest X-ray of a 29-year-old immunocompromised female patient with a 3-day history of cough and fever. Past medical history includes severe ulcerative colitis treated with tofacitinib. b Chest X-ray of case number 59,554 presented to emergency department with respiratory distress and dyspnea. c Chest X-ray of the case number 56,442 with a history of shortness of breath and cough, and recent international travel with someone who tested positive for COVID-19. d CT scan of the case number 56,442
Discussion
Our investigations use state-of-the-art benchmark SSL methods for the potential diagnosis of COVID using X-ray and CT-Scan for the first time. We observe that COVIDCon exhibits significantly improved performance compared to other state-of-the-art methods in all settings that we have studied. For limited labeled data, COVIDCon performs exceptionally well. For instance, at 1000 labels, on the COVID-19 Radiography Dataset, it achieves an accuracy of 97.07%, which is 7.65% better than the next best VAT model [40]. The improved performance of COVIDCon on such a small dataset signifies its use in the medical diagnosis domain, where it is often difficult to obtain large annotated datasets. COVIDCon also works extremely well on a larger COVID-19 CT Scan Dataset. It achieves an accuracy of 99.13%, which is 6.5% superior to its closest competitor, Pseudo-label. These results demonstrate COVIDCon as the benchmark SSL algorithm for potential diagnosis of COVID-19 from chest X-rays and CT-Scans.
Furthermore, COVIDCon performs exceptionally well in identifying COVID-19 positive cases from completely unseen chest X-rays and CT scans; therefore has potentials to be used in clinical settings. In that context, additional attributes, such as demographic information, race, etc. can also be included in COVIDCon for strengthening the ground for classification. It will be also important to understand the possibilities of identifying asymptomatic COVID-19 cases and differentiate them from normal cases from X-rays and CT scans screening using COVIDCon. For that, a large amount of data from normal, asymptomatic, as well as symptomatic COVID-19 cases will be required and collaborative work between hospitals and machine learning scientists will be necessary. Future studies will be focused on making the proposed approach available to the society for use in early and efficient detection of COVID-19.
Data availability
All the data and materials are available on request to the authors (L.K., P.S.). We have made the codes available online. Please refer to the github link https://github.com/HiPracheta/COVIDCon
Notes
References
Abbas A, Abdelsamea MM, Gaber MM (2021) Classification of covid-19 in chest X-ray images using detrac deep convolutional neural network. Appl Intell 51(2):854–864
Abdel-Basset M, Chang V, Hawash H, Chakrabortty RK, Ryan M (2021) FSS-2019-NCOV: a deep learning architecture for semi-supervised few-shot segmentation of covid-19 infection. Knowl-Based Syst 212:106647
Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 296:32–40
Anand A, Racah E, Ozair S, Bengio Y, Côté MA, Hjelm RD (2019) Unsupervised state representation learning in atari. arXiv:1906.08226
Apostolopoulos ID, Mpesiana TA (2020) Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med 43(2):635–640
Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A (2020) Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med 121:103795
Axell-House DB, Lavingia R, Rafferty M, Clark E, Amirian ES, Chiao EY (2020) The estimation of diagnostic accuracy of tests for COVID-19: a scoping review. J Infect 81:681–697
Bachman P, Alsharif O, Precup D (2014) Learning with pseudo-ensembles. Adv Neural Inf Process Syst 27:3365–3373
Bachman P, Hjelm RD, Buchwalter W (2019) Learning representations by maximizing mutual information across views. arXiv:1906.00910
Berthelot D, Carlini N, Goodfellow I, Papernot N, Oliver A, Raffel CA (2019) Mixmatch: a holistic approach to semi-supervised learning. In: NIPS, pp 5050–5060
Caruana R, Niculescu-Mizil A (2006) An empirical comparison of supervised learning algorithms. In Proceedings of the 23rd international conference on Machine learning, pp 161–168
Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al Emadi N et al (2020) Can AI help in screening viral and covid-19 pneumonia? IEEE Access 8:132665–132676
Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection. arXiv:2003.11597
Das D, Santosh K, Pal U (2020) Truncated inception net: Covid-19 outbreak screening using chest X-rays. Phys Eng Sci Med 43(3):915–925
El Asnaoui K, Chawki Y, Idri A (2021) Automated methods for detection and classification pneumonia based on X-ray images using deep learning. Artificial intelligence and blockchain for future cybersecurity applications. Springer, New York, pp 257–284
Esteva A, Kuprel B, Novoa RA et al (2017) Correction: Corrigendum: Dermatologist-level classification of skin cancer with deep neural networks. Nature 546(7660):686–686
Fan DP, Zhou T, Ji GP, Zhou Y, Chen G, Fu H, Shen J, Shao L (2020) Inf-net: Automatic covid-19 lung infection segmentation from CT images. IEEE Trans Med Imaging 39(8):2626–2637
He K, Fan H, Wu Y, Xie S, Girshick R (2020) Momentum contrast for unsupervised visual representation learning. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 9729–9738
Hemdan EED, Shouman MA, Karar ME (2020) Covidx-net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv:2003.11055
Henaff O (2020) Data-efficient image recognition with contrastive predictive coding. In International conference on machine learning, pp. 4182–4192. PMLR
Hjelm RD, Fedorov A, Lavoie-Marchildon S, Grewal K, Bachman P, Trischler A, Bengio Y (2018) Learning deep representations by mutual information estimation and maximization. arXiv:1808.06670
Islam MM, Karray F, Alhajj R, Zeng J (2021) A review on deep learning techniques for the diagnosis of novel coronavirus (covid-19). IEEE Access 9:30551–30572. https://doi.org/10.1109/ACCESS.2021.3058537
Ismael AM, Şengür A (2021) Deep learning approaches for covid-19 detection based on chest X-ray images. Expert Syst Appl 164:114054
Jain R, Gupta M, Taneja S, Hemanth DJ (2021) Deep learning based detection and analysis of covid-19 on chest X-ray images. Appl Intell 51(3):1690–1700
Jin C, Chen W, Cao Y, Xu Z, Tan Z, Zhang X, Deng L, Zheng C, Zhou J, Shi H et al (2020) Development and evaluation of an artificial intelligence system for covid-19 diagnosis. Nat Commun 11(1):1–14
Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH (2020) Essentials for radiologists on COVID-19: an update–radiology scientific expert panel. Radiology 296:2
Khobahi S, Agarwal C, Soltanalian M (2020) Coronet: a deep network architecture for semi-supervised task-based identification of covid-19 from chest x-ray images. MedRxiv 17:1–16
Konar D, Panigrahi BK, Bhattacharyya S, Dey N, Jiang R (2021) Auto-diagnosis of covid-19 using lung CT images with semi-supervised shallow learning network. IEEE Access 9:28716–28728
Kubista M, Andrade JM, Bengtsson M, Forootan A et al (2006) The real-time polymerase chain reaction. Mol Asp Med 27(2–3):95–125
Kurakin A, Raffel C, Berthelot D, Cubuk ED, Zhang H, Sohn K, Carlini N (2020) Remixmatch: semi-supervised learning with distribution matching and augmentation anchoring
Laine S, Aila T (2016) Temporal ensembling for semi-supervised learning. arXiv:1610.02242
Lee DH (2013) Pseudo-label: The simple and efficient semi-supervised learning method for deep neural networks. In: Workshop on challenges in representation learning, ICML, vol. 3, p 2
Lee EY, Ng MY, Khong PL (2020) COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis 20(4):384–385
Li R, Zhang W, Suk HI, Wang L, Li J, Shen D, Ji S (2014) Deep learning based imaging data completion for improved brain disease diagnosis. In International conference on medical image computing and computer-assisted intervention. Springer, New York pp 305–312
Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q et al (2020) Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest CT. Radiology
Ma J, Nie Z, Wang C, Dong G, Zhu Q, He J, Gui L, Yang X (2020) Active contour regularized semi-supervised learning for covid-19 CT infection segmentation with limited annotations. Phys Med Biol 65(22):225034
Maaten LVD, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9:2579–2605
Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Mirjalili S, Khan MK (2021) Diagnosing covid-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. Int Soc Opt Photon 11734:117340E
McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, Back T, Chesus M, Corrado GC, Darzi A et al (2020) International evaluation of an AI system for breast cancer screening. Nature 577(7788):89–94
Miyato T, Si Maeda, Koyama M, Ishii S (2018) Virtual adversarial training: a regularization method for supervised and semi-supervised learning. TPAMI 41(8):1979–1993
Narin A, Kaya C, Pamuk Z (2021) Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks. Pattern Anal Appl 57:1–14
Nour M, Cömert Z, Polat K (2020) A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl Soft Comput 97:106580
Oord Avd, Li Y, Vinyals O (2018) Representation learning with contrastive predictive coding. arXiv:1807.03748
Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR (2020) Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med 121:103792
Panwar H, Gupta P, Siddiqui MK, Morales-Menendez R, Singh V (2020) Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos Solit Fractals 138:109944
Pathak Y, Shukla PK, Tiwari A, Stalin S, Singh S, Shukla PK (2020) Deep transfer learning based classification model for COVID-19 disease. IRBM
Rahman T, et al (2020) COVID-19 radiography database. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database
Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, Lungren MP, Ng AY (2017) Chexnet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv:1711.05225
Sahoo P, Roy I, Wang Z, Mi F, Yu L, Balasubramani P, Khan L, Stoddart JF (2020) Multicon: a semi-supervised approach for predicting drug function from chemical structure analysis. J Chem Inf Model 60:5995–6006
Sajjadi M, Javanmardi M, Tasdizen T (2016) Mutual exclusivity loss for semi-supervised deep learning. In 2016 IEEE International Conference on Image Processing (ICIP), pp 1908–1912. IEEE
Shankar K, Perumal E (2021) A novel hand-crafted with deep learning features based fusion model for covid-19 diagnosis and classification using chest x-ray images. Complex Intell Syst 7(3):1277–1293
shensheng Xu S, Mak MW, Cheung CC (2017) Deep neural networks versus support vector machines for ECG arrhythmia classification. In: 2017 IEEE international conference on multimedia & expo workshops (ICMEW), pp 127–132. IEEE
Sohn K, Berthelot D, Li CL, Zhang Z, Carlini N, Cubuk ED, Kurakin A, Zhang H, Raffel C (2020) Fixmatch: simplifying semi-supervised learning with consistency and confidence. arXiv:2001.07685
Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Wang R, Zhao H, Zha Y et al (2021) Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with CT images. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2021.3065361
Tarvainen A, Valpola H (2017) Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. In: Advances in neural information processing systems, pp 1195–1204
Tian Y, Krishnan D, Isola P (2020) Contrastive multiview coding. In: Computer vision–ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part XI 16, pp 776–794. Springer
Toğaçar M, Ergen B, Cömert Z (2020) COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med 121:103805
Ucar F, Korkmaz D (2020) Covidiagnosis-net: deep Bayes-squeezenet based diagnostic of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses 140:109761
Velickovic P, Fedus W, Hamilton WL, Liò P, Bengio Y, Hjelm RD (2019) Deep graph infomax. In: ICLR (Poster)
Verma V, Lamb A, Kannala J, Bengio Y, Lopez-Paz D (2019) Interpolation consistency training for semi-supervised learning. IJCAI
Wang L, Lin ZQ, Wong A (2020) Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest X-ray images. Sci Rep 10(1):1–12
Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X et al (2021) A deep learning algorithm using CT images to screen for corona virus disease (covid-19). Eur Radiol 16:1–9
Wehbe RM, Sheng J, Dutta S, Chai S, Dravid A, Barutcu S, Wu Y, Cantrell DR, Xiao N, Allen BD et al (2020) Deepcovid-xr: an artificial intelligence algorithm to detect COVID-19 on chest radiographs trained and tested on a large us clinical dataset. Radiology 299:167
Whitaker M, Kron T, Sobolewski M, Dove R (2020) Covid-19 pandemic planning: considerations for radiation oncology medical physics. Phys Eng Sci Med 43(2):473–480
Xie S, Girshick R, He K et al (2017) Aggregated residual transformations for deep neural networks. In: CVPR 1492–1500
Xie Q, Dai Z, Hovy E, Luong MT, Le QV (2019) Unsupervised data augmentation for consistency training. arXiv:1904.12848
Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical 2019-ncov pneumonia: relationship to negative RT-PCR testing. Radiology 296:41–45
Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J et al (2020) A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering 6(10):1122–1129
Yi D, Lei Z, Liao S, Li SZ (2014) Deep metric learning for person re-identification. In: 2014 22nd international conference on pattern recognition, pp 34–39. IEEE
Yoon SH, Lee KH, Kim JY, Lee YK, Ko H, Kim KH, Park CM, Kim YH (2020) Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): analysis of nine patients treated in korea. Korean J Radiol 21(4):494–500
Zhang K, Liu X, Shen J, Li Z, Sang Y, Wu X, Zha Y, Liang W, Wang C, Wang K et al (2020) Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell 181:1423
Zhao J, Zhang Y, He X, Xie P (2020) COVID-CT-dataset: a CT scan dataset about COVID-19. arXiv:2003.13865
Zhu X, Goldberg AB (2009) Introduction to semi-supervised learning. Synth Lect Artif Intell Mach Learn 3(1):1–130
Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ (2020) Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology 296:200490
Acknowledgements
(Pracheta Sahoo and Indranil Roy contributed equally to this work). The authors thank Zhuoyi Wang for his valuable insights and support.
Funding
The research reported herein was supported in part by NSF Awards (Grant Nos. DMS-1737978, DGE-2039542, OAC1828467, OAC-1931541, DGE-1906630) and an IBM Faculty Award (Research).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declared no conflict of interest.
Code availability
Not applicable.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
The authors give consent to publish the paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sahoo, P., Roy, I., Ahlawat, R. et al. Potential diagnosis of COVID-19 from chest X-ray and CT findings using semi-supervised learning. Phys Eng Sci Med 45, 31–42 (2022). https://doi.org/10.1007/s13246-021-01075-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-021-01075-2