Abstract
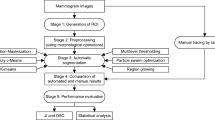
The presented study aims to design a computer-aided detection and diagnosis system for breast dynamic contrast enhanced magnetic resonance imaging. In the proposed system, the segmentation task is performed in two stages. The first stage is called breast region segmentation in which adaptive noise filtering, local adaptive thresholding, connected component analysis, integral of horizontal projection, and breast region of interest detection algorithms are applied to the breast images consecutively. The second stage of segmentation is breast lesion detection that consists of 32-class Otsu thresholding and Markov random field techniques. Histogram, gray level co-occurrence matrix and neighboring gray tone difference matrix based feature extraction, Fisher score based feature selection and, tenfold and leave-one-out cross-validation steps are carried out after segmentation to increase the reliability of the designed system while decreasing the computational time. Finally, support vector machines, k- nearest neighbor, and artificial neural network classifiers are performed to separate the breast lesions as benign and malignant. The average accuracy, sensitivity, specificity, and positive predictive values of each classifier are calculated and the best results are compared with the existing similar studies. According to the achieved results, the proposed decision support system for breast lesion segmentation distinguishes the breast lesions with 86%, 100%, 67%, and 85% accuracy, sensitivity, specificity, and positive predictive values, respectively. These results show that the proposed system can be used to support the radiologists during a breast cancer diagnosis.










Similar content being viewed by others
References
Lee CI, Lehman CD, Bassett LW (2018) Breast imaging. Oxford University Press, Oxford
World Health Organization (2019) https://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/. Accessed 5 Mar 2019
American Institute for Cancer Research Breast Cancer Statistics (2014) https://www.wcrf.org/dietandcancer/cancer-trends/breast-cancer-statistics. Accessed 5 Mar 2019
T. C. Ministry of Health Public Health Agency of Turkey, Turkey Cancer Statistics (2019) https://hsgm.saglik.gov.tr/depo/birimler/kanser-db/istatistik/2014-RAPOR._uzun.pdf. Accessed 5 Mar 2019
Losurdo L, Fanizzi A, Basile TM, Bellotti R, Bottigli U, Dentamaro R, et al. (2018) A combined approach of multiscale texture analysis and interest point/corner detectors for microcalcifications diagnosis. In: International conference on bioinformatics and biomedical engineering. Springer, Cham, pp 302–313. https://doi.org/10.1007/978-3-319-78723-7_26
Pandey D, Yin X, Wang H, Su MY, Chen JH, Wu J, Zhang Y (2018) Automatic and fast segmentation of breast region-of-interest (ROI) and density in MRIs. Heliyon 4:1–30. https://doi.org/10.1016/j.heliyon.2018.e01042
Illan IA, Ramirez J, Gorriz JM (2018) Automated detection and segmentation of nonmass-enhancing breast tumors with dynamic contrast-enhanced magnetic resonance imaging. Contrast Media Mol Imaging. https://doi.org/10.1155/2018/5308517
Shokouhi SB, Fooladivanda A, Ahmadinejad N (2017) Computer-aided detection of breast lesions in DCE-MRI using region growing based on fuzzy C-means clustering and vesselness filter. Eurasip J Adv Sig Process. https://doi.org/10.1186/s13634-017-0476-x
Marrone S, Piantadosi G, Fusco R, Petrillo A, Sansone M, Sansone C (2016) Breast segmentation using Fuzzy C-Means and anatomical priors in DCE-MRI. In: Proceedings of the international conference on pattern recognition, pp 1472–1477. https://doi.org/10.1109/ICPR.2016.7899845
Waugh SA, Purdie CA, Jordan LB, Vinnicombe S, Lerski RA (2016) Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol 26:322–330. https://doi.org/10.1007/s00330-015-3845-6
Tzalavra A, Dalakleidi K, Zacharaki EI, Tsiaparos N (2016) Comparison of multi-resolution analysis patterns for texture classification of breast tumors based on DCE-MRI. In: 7th international workshop on machine learning in medical imaging (MICCAI), pp 296–304. https://doi.org/10.1007/978-3-319-47157-0_36
Honda E, Nakayama R, Koyama H, Yamashita A (2016) Computer-aided diagnosis scheme for distinguishing between benign and malignant masses in breast DCE-MRI. J Digit Imaging 29:388–393. https://doi.org/10.1007/s10278-015-9856-7
Antropova N, Huynh B, Giger M (2016) Predicting breast cancer malignancy on DCE-MRI data using pre-trained convolutional neural networks. Med Phys 43:3349–3350. https://doi.org/10.1118/1.4955674
Mahrooghy M, Ashraf AB (2015) Pharmacokinetic tumor heterogeneity as a prognostic biomarker for classifying breast cancer recurrence risk. IEEE Trans Biomed Eng 62:1585–1594. https://doi.org/10.1109/TBME.2015.2395812
Commons C, License A (2015) Automatic segmentation in breast cancer using watershed algorithm. Int J Biomed Eng Sci 2:1–6
Yang Q, Li L, Zheng B (2015) A new quantitative image analysis method for improving breast cancer diagnosis using DCE-MRI examinations. Med Phys 42:103–109. https://doi.org/10.1118/1.4903280
Navaei-Lavasani S, Fathi-Kazerooni A, Saligheh-Rad H, Gity M (2015) Discrimination of benign and malignant suspicious breast tumors based on semi-quantitative DCE-MRI parameters employing support vector machine. Front Biomed Technol 2:87–92
Chaudhurya B, Zhou M, Goldgof DB, Hall LO (2015) Identifying metastatic breast tumors using textural kinetic features of a contrast based habitat in DCE-MRI. Med Imaging. https://doi.org/10.1117/12.2081386
Hassanien AE, Moftah H, Azar AT, Shoman M (2014) MRI breast cancer diagnosis hybrid approach using adaptive ant-based segmentation and multilayer perceptron neural networks classifier. Appl Soft Comput J 14:62–71. https://doi.org/10.1016/j.asoc.2013.08.011
Song H, Zhang Q, Sun F, Wang J (2014) Breast tissue segmentation on MR images using KFCM with spatial constraints. In: Proceedings of the 2014 IEEE international conference on granular computing GrC, pp 254–258. https://doi.org/10.1109/GRC.2014.6982845
Al-faris AQ, Ngah UK, Ashidi N, Isa M, Shuaib IL (2014) Breast MRI tumour segmentation using modified automatic seeded region growing based on particle swarm optimization image clustering. Soft Comput Ind Appl 223:49–60
Wang TC, Huang YH, Huang CS, Chen JH (2014) Computer-aided diagnosis of breast DCE-MRI using pharmacokinetic model and 3-D morphology analysis. Magn Reson Imaging 32:197–205. https://doi.org/10.1016/j.mri.2013.12.002
Jayender J, Chikarmane S, Jolesz FA, Gombos E (2014) Automatic segmentation of invasive breast carcinomas from dynamic contrast-enhanced MRI using time series analysis. J Magn Reson Imaging 40:467–475. https://doi.org/10.1002/jmri.24394
Cai H, Peng Y, Ou C, Chen M, Li L (2014) Diagnosis of breast masses from dynamic contrast-enhanced and diffusion-weighted MR: a machine learning approach. PLoS ONE 9:873–887. https://doi.org/10.1371/journal.pone.0087387
Chen JH, Chen S, Chan S, Lin M, Su MY, Wang X (2013) Template-based automatic breast segmentation on MRI by excluding the chest region. Med Phys 40:22301. https://doi.org/10.1118/1.4828837
Yang Q, Li L, Zhang J, Shao G, Zhang C, Zheng B (2013) Computer-aided diagnosis of breast DCE-MRI images using bilateral asymmetry of contrast enhancement between two breasts. J Digit Imaging 27:152–160. https://doi.org/10.1007/s10278-013-9617-4
Wu S, Weinstein SP, Conant EF, Schnall MD, Kontos D (2013) Automated chest wall line detection for whole-breast segmentation in sagittal breast MR images. Med Phys 40:042301. https://doi.org/10.1118/1.4793255
Wang Y, Morrell G, Heibrun ME, Payne A, Parker DL (2013) 3D multi-parametric breast MRI segmentation using hierarchical support vector machine with coil sensitivity correction. Acad Radiol 20:137–147. https://doi.org/10.1016/j.acra.2012.08.016
Sathya DJ, Geetha K (2013) Experimental investigation of classification algorithms for predicting lesion type on breast DCE-MR images. Int J Comput Appl 82:1–8. https://doi.org/10.5120/14101-2125
Sathya DJ, Geetha K (2013) Mass classification in breast DCE-MR images using an artificial neural network trained via a bee colony optimization algorithm. Sci Asia 39:294–305. https://doi.org/10.2306/scienceasia1513-1874.2013.39.294
Nagarajan MB, Huber MB, Schlossbauer T, Leinsinger G, Krol A, Wismüller A (2013) Classification of small lesions in dynamic breast MRI: eliminating the need for precise lesion segmentation through spatio-temporal analysis of contrast enhancement. Mach Vis Appl 24:1371–1381. https://doi.org/10.1007/s00138-012-0456-y
McClymont D, Trakic A, Mehnert A, Crozier S, Kennedy D (2013) Fully automatic lesion segmentation in breast MRI using mean-shift and graph-cuts on a region adjacency graph. J Magn Reson Imaging 39:795–804. https://doi.org/10.1002/jmri.24229
Glaßer S, Niemann U, Preim B, Spiliopoulou M (2013) Can we distinguish between benign and malignant breast tumors in DCE-MRI by studying a tumor’s most suspect region only? In: Proceedings of 26th IEEE international symposium of computing medical system, pp 77–82. https://doi.org/10.1109/CBMS.2013.6627768
Wang L, Platel B, Ivanovskaya T, Harz M, Hahn HK (2012) Fully automatic breast segmentation in 3D breast MRI. In: Proceedings of international symposium of biomedical imaging, pp 1024–1027. https://doi.org/10.1109/ISBI.2012.6235732
Nagarajan MB, Huber MB, Schlossbauer T, Leinsinger G, Krol A, Wismüller A (2012) Classification of small lesions in breast MRI: evaluating the role of dynamically extracted texture features through feature selection. J Med Biol Eng 33:59–68. https://doi.org/10.5405/jmbe.1183
Hassanien AE, Kim TH (2012) Breast cancer MRI diagnosis approach using support vector machine and pulse coupled neural networks. J Appl Log 10:277–284. https://doi.org/10.1016/j.jal.2012.07.003
Fusco R, Sansone M, Petrillo A, Sansone C (2012) A multiple classifier system for classification of breast lesions using dynamic and morphological features in DCE-MRI. Lect Notes Comput Sci 7626:684–692
Vignati A, Giannini V, De Luca M, Voral L (2011) Performance of a fully automatic lesion detection system for breast DCE-MRI. J Magn Reson Imaging 34:1341–1351. https://doi.org/10.1002/jmri.22680
Kannan SR, Ramathilagam S, Devi R, Sathya A (2011) Robust kernel FCM in segmentation of breast medical images. Expert Syst Appl 38:4382–4389. https://doi.org/10.1016/j.eswa.2010.09.107
Agner SC, Soman S, Libfeld E, McDonald M (2011) Textural kinetics: a novel dynamic contrast-enhanced (DCE)-MRI feature for breast lesion classification. J Digit Imaging 24:446–463. https://doi.org/10.1007/s10278-010-9298-1
Giannini V, Vignati A, Morra L, Persano D (2010) A fully automatic algorithm for segmentation of the breasts in DCE-MR images. In: Annual international conferences of the IEEE engineering in medicine and biology society in EMBC, pp 3146–3149. https://doi.org/10.1109/IEMBS.2010.5627191
Lee SH, Kim JH, Cho N, Yang Z (2010) Multilevel analysis of spatiotemporal association features for differentiation of tumor enhancement patterns in breast DCE-MRI. Med Phys 37:3940–3956. https://doi.org/10.1118/1.3446799
Dyck DV, Backer SD, Juntu J, Sijbers J, Rajan J (2010) Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Imaging 31:680–689. https://doi.org/10.1002/jmri.22095
Newstead GM, Lan L, Giger ML, Li H, Jansen SA, Bhooshan N (2010) Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology 254:680–690. https://doi.org/10.1148/radiol.09090838
Manuscript A, Levman J, Leung T, Causer P, Martel AL (2010) Resonance breast lesions by support vector machines. IEEE Trans Med Imaging 27:688–696. https://doi.org/10.1109/TMI.2008.916959
Zheng Y, Englander S (2009) STEP: Spatiotemporal enhancement pattern for MR-based breast tumor diagnosis. Med Phys 36:3192–3204. https://doi.org/10.1118/1.3151811
Newell D, Nie K, Chen JH, Hsu CC (2009) Selection of diagnostic features on breast MRI to differentiate between malignant and benign lesions using computer-aided diagnosis: differences in lesions presenting as mass and non-mass-like enhancement. Eur Radiol 20:771–781. https://doi.org/10.1007/s00330-009-1616-y
McLaren CE, Chen WP, Nie K, Su MY (2009) Prediction of malignant breast lesions from MRI features: a comparison of artificial neural network. Acad Radiol 16:842–851. https://doi.org/10.1016/j.acra.2009.01.029
Lee SH, Kim JH, Park JS, Jung YS, Moon WK (2009) Characterizing time-intensity curves for spectral morphometric analysis of intratumoral enhancement patterns in breast DCE-MRI: comparison between differentiation performance of temporal model parameters based on DFT AND SVD. In: IEEE international symposium on biomedical imaging: from nano to macro, ISBI 2009, pp 65–68. https://doi.org/10.1109/ISBI.2009.5192984
Furman-Haran E, Eyal E, Kirshenbaum KJ, Kelcz F, Degani H, Badikhi D (2009) Principal component analysis of breast DCE-MRI adjusted with a model-based method. J Magn Reson Imaging 30:989–998. https://doi.org/10.1002/jmri.21950
Agner SC, Xu J, Fatakdavala H, Ganesan S (2009) Segmentation and classification of triple negative breast cancers using DCE-MRI. In: Proceedings of 2009 IEEE international symposium on biomedical imaging: from nano to macro, ISBI 2009, pp 1227–1230. https://doi.org/10.1109/ISBI.2009.5193283
Twellmann T, Romeny BT (2008) Computer-aided classification of lesions by means of their kinetic signatures in dynamic contrast-enhanced MR images. Proc SPIE. https://doi.org/10.1117/12.769651
Mayerhoefer ME, Breitenseher M, Amann G, Dominkus M (2008) Are signal intensity and homogeneity useful parameters for distinguishing between benign and malignant soft tissue masses on MR images. Magn Reson Imaging 26:1316–1322. https://doi.org/10.1016/j.mri.2008.02.013
Lee SH, Kim JH, Park JS, Chang CM (2008) Computerized segmentation and classification of breast lesions using perfusion volume fractions in dynamic contrast-enhanced MRI. Biomed Eng Inform New Dev Futur 2:58–62. https://doi.org/10.1109/BMEI.2008.215
Ertaş G, Gülçür HÖ, Osman O, Uçan ON, Tunaci M, Dursun M (2008) Breast MR segmentation and lesion detection with cellular neural networks and 3D template matching. Comput Biol Med 38:116–126. https://doi.org/10.1016/j.compbiomed.2007.08.001
Woods BJ, Clymer BD, Kurc T, Heverhagen JT (2007) Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data. J Magn Reson Imaging 25:495–501. https://doi.org/10.1002/jmri.20837
Meinel LA, Stolpen AH, Berbaum KS, Fajardo LL, Reinhardt JM (2007) Breast MRI lesion classification: improved performance of human readers with a backpropagation neural network computer-aided diagnosis (CAD) system. J Magn Reson Imaging 25:89–95. https://doi.org/10.1002/jmri.20794
Khazen M, Leach MO, Hawkes DJ, Tanner C, Kessar P (2006) Does registration improve the performance of a computer aided diagnosis system for dynamic contrast-enhanced MR mammography? In: 3rd IEEE international symposium biomedical imaging nano to macro, pp 466–469. https://doi.org/10.1109/ISBI.2006.1624954
Leinsinger G, Schlossbauer T, Scherr M, Lange O, Reiser M, Wismüller A (2006) Cluster analysis of signal-intensity time course in dynamic breast MRI: does unsupervised vector quantization help to evaluate small mammographic lesions? Eur Radiol 16:1138–1146. https://doi.org/10.1007/s00330-005-0053-9
Chen W, Giger ML, Bick U, Newstead GM (2006) Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI. Med Phys 33:2878–2887. https://doi.org/10.1118/1.2210568
Xiaohua C, Brady M, Lo JJ (2005) Simultaneous segmentation and registration of contrast-enhanced breast MRI. Inf Process Med Imaging 3565:31–59
Arbach L, Stolpen A, Reinhardt JM (2004) Classification of breast MRI lesions using a backpropagation neural network (BNN). In: IEEE international symposium on biomedical imaging nano to macro (IEEE cat no. 04EX821), pp 253–256. https://doi.org/10.1109/ISBI.2004.1398522
Vomweg TW, Buscema M, Kauczor HU, Teifke A (2003) Improved artificial neural networks in prediction of malignancy of lesions in contrast-enhanced MR-mammography. Med Phys 30:2350–2359. https://doi.org/10.1118/1.1600871
Tzacheva AA, Najarian K, Brockway JP (2003) Breast cancer detection in gadolinium-enhanced MR images by static region descriptors and neural networks. J Magn Reson Imaging 17:337–342. https://doi.org/10.1002/jmri.10259
Lucht R, Delorme S, Brix G (2002) Neural network-based segmentation of dynamic MR mammographic images. Magn Res Imaging 20:147–154. https://doi.org/10.1016/S0730-725X(02)00464-2
Abdolmaleki P, Buadu LD, Naderimansh H (2001) Feature extraction and classification of breast cancer on dynamic magnetic resonance imaging using artificial neural network. Cancer Lett 171:183–191. https://doi.org/10.1016/s0304-3835(01)00508-0
Vergnaghi D, Monti A, Setti E, Musumeci R (2001) A use of a neural network to evaluate contrast enhancement curves in breast magnetic resonance images. J Digit Imaging 14:58–59. https://doi.org/10.1007/bf03190297
Deng H, Clausi DA (2004) Unsupervised image segmentation using a simple MRF model with a new implementation scheme. Pattern Recogn 37:2323–2335. https://doi.org/10.1016/j.patcog.2004.04.015
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 3:610–621. https://doi.org/10.1109/TSMC.1973.4309314
Bishop CM (1995) Neural networks for pattern recognition. Oxford University Press, Oxford
Bishop CM (2006) Pattern recognition and machine learning. BMEI, New York
Duda RO, Hart PE, Stark DG (2001) Pattern classification, 2nd edn. Wiley, New York
Haykin S (2009) Neural networks and learning machines. Prentice Hall, Upper Saddle River
Evgeniou T, Pentil M (2001) Support vector machines: theory and applications machine learning and its applications advanced lectures. Mach Learn Its Appl. https://doi.org/10.1007/3-540-44673-7_12
Piantadosi G, Marrone S, Fusco R, Sansone M, Sansone C (2018) Comprehensive computer-aided diagnosis for breast T1-weighted DCE-MRI through quantitative dynamical features and spatio-temporal local binary patterns. IET Comput Vis 12:1007–1017
Fooladivanda A, Shokouki BS, Ahmedinejad N, Mosaui MR (2015) Automatic segmentation of breast and fibroglandular tissue in MRI using local adaptive thresholding. ICMBE 2015:195–200. https://doi.org/10.1049/iet-cvi.2018.5273
Gül S, Çetinel G (2017) Detection of lesion boundaries in breast magnetic resonance imaging with OTSU thresholding and fuzzy C-means. ICAT 2017:1–5
Marrone S, Pantodosi G, Fusco R, Sansone M, Sansone C (2017) An investigation of deep learning for lesions malignancy classification in breast DCE-MRI. In: International conference on image analysis and processing, pp 479–489. https://doi.org/10.1007/978-3-319-68548-9_44
Losurdo L, Basile TMA, Fanizzi A, Bellotti R, Bottigli U, Carbonara R et al (2018) A gradient-based approach for breast DCE-MRI analysis. Biomed Res Int. https://doi.org/10.1155/2018/9032408
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors Dr. Gökçen Çetinel is the coordinator, Dr. Fuldem Mutlu is the researcher and Sevda Gül is the scholarship student in the project that is supported by The Scientific and Technical Research Council of Turkey (TUBITAK) through The Research Support Programs Directorate (ARDEB) with Project Number of 118E201.
Ethical approval
The study was approved by the Ethical Committee of Sakarya University Medical Sciences Faculty. All procedures in studies involving human participants were in accordance with the ethical standards of the institutional and/or national search committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Çetinel, G., Mutlu, F. & Gül, S. Decision support system for breast lesions via dynamic contrast enhanced magnetic resonance imaging. Phys Eng Sci Med 43, 1029–1048 (2020). https://doi.org/10.1007/s13246-020-00902-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-020-00902-2