Abstract
In crop protection, disease quantification parameters such as disease incidence (DI) and disease severity (DS) are the principal indicators for decision making, aimed at ensuring the safety and productivity of crop yield. The quantification is standardized with leaf organs, defined as individual scoring units. This study focuses on identifying and segmenting individual leaves in agricultural fields using unmanned aerial vehicle (UAV), multispectral imagery of sugar beet fields, and deep instance segmentation networks (Mask R-CNN). Five strategies for achieving network robustness with limited labeled images are tested and compared, employing simple and copy-paste image augmentation techniques. The study also evaluates the impact of environmental conditions on network performance. Metrics of performance show that multispectral UAV images recorded under sunny conditions lead to a performance drop. Focusing on the practical application, we employ Mask R-CNN models in an image-processing pipeline to calculate leaf-based parameters including DS and DI. The pipeline was applied in time-series in an experimental trial with five varieties and two fungicide strategies to illustrate epidemiological development. Disease severity calculated with the model with highest Average Precision (AP) shows the strongest correlation with the same parameter assessed by experts. The time-series development of disease severity and disease incidence demonstrates the advantages of multispectral UAV-imagery in contrasting varieties for resistance, as well as the limits for disease control measurements. This study identifies key components for automatic leaf segmentation of diseased plants using UAV imagery, such as illumination and disease condition. It also provides a tool for delivering leaf-based parameters relevant to optimize crop production through automated disease quantification by imaging tools.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Imaging sensors attached to unmanned aerial vehicles (UAVs) are systems that currently revolutionize the way to monitor agricultural fields [1]. Agricultural management practices are feasible after analysis of high spatial and temporal resolution images, allowing decision making at the right place, with the right intensity, and at the right time. The list of practical applications of UAV-systems starts with the area of plant breeding and phenotyping, and has further applications for plant protection, yield prediction, growth vigor, nutrient status, weed detection and drought stress. Moreover, UAV-systems support precision agriculture because crop production is optimized by maintaining or increasing yield, while reducing environmental impact and resources for pest, water and nutrient management [2].
Disease quantification is a common visual scoring activity for plant breeding and decision making in plant protection nowadays. This activity must be executed several times over vegetation development by appropriately trained staff or experts [3]. The most established parameters for quantification are disease incidence (DI) and disease severity (DS) [4, 5]. For phenotyping DI and DS in sugar beets, experts collect a representative number of sugar beet leaves, which are assessed for presence or absence of symptoms, and for determining the degree of damage [6]. According to Reynolds et al. [7], phenotyping work like visual scoring represents the major proportion of costs in experimental trials. Variability and repeatability of collected data is also an issue in field assessments: “Inter-rater” errors occur when different experts are involved in phenotyping at various key disease development stages. The high manpower requirement associated with this error source is a disadvantage for extensive experimental fields. Similarly, other factors negatively influence the variability and repeatability of observations, including the effect of noise, heat, exhaustion or time allocated for an assessment [5]. All those factors emphasize the limitations of visual scoring methods and motivate the development of innovative and automated UAV-based imaging approaches for quantification of plant diseases [8,9,10,11].
In the past, studies used RGB and multispectral UAV-systems for disease quantification in sugar beet fields. Jay et al. [8] and Görlich et al. [9] segment limits of plot regions from RGB and multispectral orthomosaic images to calculate DS in variety trials. Similarly, a pipeline by Günder et al. [12] to segment individual plants, was extended for an application in disease quantification of Cercospora leaf spot (CLS) to classify infested plants according to their disease categories [10]. Circular regions within a plot are considered for an automated analysis of multispectral images to calculate DI, DS, and additional parameters such as area of foliage, area of healthy foliage, number of lesions and mean area of lesion by unit of foliage [11]. Whether circular-, plant-, or plot-based regions, this image-based scoring shows robustness to calculate DS. Nevertheless, DI is an aspect to improve: Barreto et al. [11] highlight the disadvantages of delimiting scoring in circle regions within a plot. False positive pixel classification of non-diseased regions leads to inaccurate quantification of diseased units for DI. Visualization of scored multispectral images shows that the principal reason of disease misclassification is the pixel quantification of harvest residues of previous crop laying on soil regions. This highlights a potential application of leaf segmentation because image-regions with high misclassification rate are removed from determining DI. While leaf segmentation could also contribute to DS, this has not been tested yet.
Image-based leaf phenotyping requires detecting and delineating a representative number of leaves for a later parameter calculation. Sugar beet canopy is a complex structure of non-uniform leaves. Individual leaves are positioned with extreme overlap, mutual occlusion, at different heights and diverse orientation. Moreover, leaf appearance is dynamic, either by natural develop stages and senescence, or by exogenous factors like diseases. Leaves change in size, and color from green to yellow, by degradation of chlorophyll content, and later from yellow to brown when necrotic tissue dominates the canopy. At last, weather conditions play also role. UAV monitoring activities must be able to cope with cloudy and sunny sky conditions if farmers are to obtain an on-time decision. Furthermore, shaded regions caused by passive illumination have to be considered for image-based individual leaf segmentation.
To solve the task of leaf identification and segmentation from UAV-data, adequate data analysis approaches are required. This task can potentially be automated by deep learning models in computer vision, more specifically instance segmentation [13]. Unfortunately, the main limitation of deep learning models in agriculture is the need for a high number of labeled images [14]. In the context of UAV-data of agricultural fields, labeling individual leaves in real images is time intensive, making this work the bottleneck for the availability of labeled data.
In this paper, we make two main contributions: (i) we address the challenge of leaf segmentation from multispectral UAV-based images with a limited number of labeled images. We evaluate augmentation approaches including basic image manipulation and copy-paste techniques to create a data set of adequate size for training instance segmentation models (Mask R-CNN). For this specific aim, we consider recommendations as described by Kurnichov et al. [15] using a copy-paste data augmentation approach for banana plantain and arabidopsis images. We identify the best model by testing performance under diverse disease and illumination conditions. (ii) We apply our best leaf segmentation model to large orthomosaic images, and integrate Mask R-CNN into a pipeline to extend the number of parameters for disease quantification beyond the circle-based parameters. Lastly, we apply the pipeline in a variety trial and evaluate the performance by comparing expert with automated scored data of DS collected in time-series.
2 Modeling Leaf Segmentation
2.1 Field Monitoring and Methodology
The monitoring campaign took place from 2019 to 2021 in four locations [11, 16]. We use multispectral UAV-systems: a DJI Inspire 2 with a 5-channel multispectral sensor, Micasense RedEdge-M; and a DJI Matrice 210 with a the 6-channel sensor variant, Micasense Altum. To get high resolution images, flight missions were planned to deliver a ground sample distance (GSD) between 2.5 and 4.1 mm. We truncate and calibrate the raw images from digital numbers to reflectance according to Barreto et al. [11]. For the photogrammetry, we use the software Agisoft Metashape Professional for stitching of truncated multichannel images, and export the output as orthomosaic. We generate image patches to cover a field area of 1.28\(\times\)1.28 m and scale these to 512\(\times\)512 px by resampling (Fig. 1a). We use five and six channel re-scaled arrays for defining RGB composite images from reflectance values by using \(V=\max (\lambda _{BLUE}, \lambda _{GREEN}, \lambda _{RED})\), and the following formulae:
Data augmentation: Manual data labeling is a very difficult and time-consuming task, particularly for sugar beet images which have highly overlapping leaves with almost identical colors. We compare five strategies (see Fig. 1b) to generate sufficient training data while keeping the labeling efforts within reasonable bounds. First, we completely label every leaf in 100 images (\(512 \times 512\) pixels) containing healthy and diseased sugar beet plants under sunny and cloudy light conditions. In total, the images contain 2951 leaves without occlusion and we split the individual leaves 80/20 into training and validation set. Eighteen additional completely labeled images serve as a hold-out test set to evaluate the performance of different models. For the background, we select 758 background UAV-images (without sugar beet plants), containing different field conditions such as: diverse soil humidity, weed pressure and soil tillage.
We use the training data and background images in four of five data augmentation strategies (I–V). For all strategies the data sets contain 10,000 images (training to validation ratio of 8000 to 2000). For strategy I we use a copy-paste augmentation technique. For this, we paste 70–140 randomly selected segmented leaves onto randomly selected background images. We use image flipping, scaling and brightness shift to transform each leave image before pasting it to the background. Likelihood and transformation factor are specified in Table 1. Strategies II to IV are mixed strategies. We combine the copy-paste method described above with the inclusion of 100 fully labeled images. Additionally, we include 100 fully labeled images and apply basic manipulation methods, such as flipping, rotation, channel shift, cropping, and brightness adjustment (Table 1). For strategy II, we use the 100 original images and created 4900 copy-paste images. Subsequently, we augment the whole set two times with simple augmentations to obtain 10,000 images. Strategy III uses the 100 original images along with 1900 copy-paste images. We augment this dataset with basic transformations five times. In the case of Strategy IV, we employ 100 original images and 100 copy-paste images, subjecting them to fifty rounds of basic augmentations. The last strategy (V) relies solely on basic manipulation, with each original image augmented 100 times.
Training: For instance segmentation, we use a Mask R-CNN architecture [17] with a ResNet-101 backbone for ROI (region of interest) prediction. We train the models on four RTX 5000 GPUs running in parallel with two images per batch (batch size = eight images) and use the adapted Mask R-CNN implementation for Tensorflow 2.0 [17] with modules Cudnn 7.6.5.32\(-\)10.2, cuda 10.1.243 and Tensorflow 2.2.0. Before training, we specify the classes “leaf” and “background”. We put the number of ROIs to train per image from 200 up to 256 and anchors to train per image from 256 up 512. We initialize our models with weights pretrained on the COCO data set [13]. Training the model for one step takes about 22 s. Since we make 500 steps per epoch we get a training time of three hours for one epoch. Mask R-CNN implements a multi-task loss, \(\mathcal {L} = \mathcal {L}_{cls} + \mathcal {L}_{box} + \mathcal {L}_{mask}\), to balance correct class predictions, accurate bounding boxes and exact masks. In this study, we trained all models for 50 epochs based on the available computational resources. During the first 25 epochs, the learning rate is kept at 0.001, then changed to 0.0001 until the end of the training.
Evaluation and metrics: We group test set images in six categories based on illumination conditions (cloudy and sunny), as well as the degree of disease damage (healthy, medium and severe). We evaluate the performance of the models with precision, recall and average precision (AP). Precision measures how many of our predicted leaves are actually leaves, while recall evaluates how many of the actual true leaves are detected by the model. Precision and recall depend on a threshold measuring intersection over union (IoU) between a predicted and a ground truth leave. We set this threshold to 50% for precision and recall. This means that to accept a prediction as correct it needs to overlap at least to 50% with a ground truth leave. The second hyperparameter is the confidence score threshold describing how confident the model must be to use the leave prediction. In this paper, we compare values of 0.5 and 0.75 for the confidence score (CS).
For AP, we measure precision and recall for all possible confidence thresholds and plot them in a curve. The area under this curve is the AP. It is large when the true predictions come with high confidence values and there are few false positives.
2.2 Instance Leaf Segmentation
Augmentation for leaf segmentation: In this section we compare the five augmentation strategies for leaf segmentation in diseased sugar beet plants. In Table 2 the AP, precision and recall under two confidence scores CSs are compared. In all approaches, by increasing the CS value, the number of false negative or missed leaves will increase. In contrast to recall, precision increases with higher CS. Increasing CS from 0.50 to 0.75 yields between 6% to 12% better precision results. The advantages of models with CS of 0.50 was reflected on a higher AP in comparison to models with 0.75 CS. Overall, the best AP was found in the model of strategy V with a value of 0.31. This strategy consist of augmenting fully labeled images only using simple techniques to increase the train and validation set to 10,000 images. Considering the study of Kuznichov et al. [15], and our findings based on the precision values obtained from strategy I in Table 2, we confirm that copy-paste augmentation strategy can serve as alternative for training Mask RCNN models for leaf segmentation. However, it is worth noting that the most efficient approach might be achieved by employing basic augmentation techniques. A reason for this behavior can be potentially attributed to the number of leaves and their relatively low resolution, which characteristics of small object detection and segmentation tasks. Two-stage detectors such as Mask R-CNN have been observed to exhibit a 50% drop in detection performance for small objects compared to larger ones [18, 19]. Difficulty to distinguish object from background, lack of well-trained features due to limited object information (number of pixels), object occlusion and truncation, are the principal challenges of two-stage detectors for quantifying small objects [18]. At this stage, when limited amount of information is a constraining factor, image context becomes relevant. Visual objects often appear within specific environments and may coexist with related objects [20]. The incorporation of contextual information in the datasets gradually increases from strategy I to V (Table 2), consequently enhancing the AP scores. The epidemiological development of the leaf disease and the nadir UAV-perspective by imaging should be an example of image context for the used data augmentation techniques, because specific environmental conditions and coexist with other related objects are not emulated. A previous study demonstrated that enhancing the performance of small object detection is attainable by incorporating additional image information (1 mm GSD) and utilizing simplified conditions. More specifically, when recording images of sugar beet plants prior to canopy closure and in the absence of disease pressure, with fewer leaves, no overlapping, and no leaf senescence, the use of Mask R-CNN yielded an AP value of 0.413 [21].
Environmental conditions for segmenting diseased leaves: We test on data sets with categories “cloudy” and “sunny” separately to further explain the performance of a Mask R-CNN leaf segmentation model under two different environmental illumination conditions. For this evaluation, the best model in terms of AP (strategy V) was analyzed in Table 3 for all six categories. Data recorded under sunny illumination conditions is the principal source for decrease of performance. UAV-images of sunny days drop in AP of 7% against cloudy and diffuse illumination. However, the question still remains as to whether the drop in performance is primarily influenced by false positive or false negative detections. A more detailed analysis of additional metrics, such as precision, reveals that this parameter experiences a more significant decrease in performance compared to the AP, while the recall remains relatively constant (resulting in a 12.3% decrease in precision). We conclude that analyzing images recorded under sunny illumination conditions increases the number of false positives or objects incorrectly detected as leaves. Illumination is an image condition that affects intra-class variation. This variation drastically impacts the performance of object detection deep learning approaches, because object appearance change in brightness and shading [22]. Furthermore, the effect of degree of disease damage is not clearly visible (Table 3). Future research will need to evaluate this last phenomena seeking the practical application of segmentation. Additionally, apart from the RGB composite, the removed multispectral channels (\(\lambda _{REDEDGE}\), \(\lambda _{NIR}\), and \(\lambda _{LWIR}\)) from original multispectral orthomosaics needs to be evaluated within alternative representations for false color composite images to determine possible improvement of leaf instance segmentation of infected fields. Finally, with the increased availability of computational resources for the training step, it is necessary to evaluate early stopping for achieving optimal leaf instance segmentation results.
3 Application of Leaf Segmentation for Parameter Extraction
3.1 Methodology for Disease Quantification in Variety Trial
We explore the potential of leaf segmentation for monitoring field experiments in a variety trial in 2021 near Göttingen, Germany. The goal of this experiment was to quantify resistance against the disease Cercospora leaf spot (CLS). We arranged a plot-trial with five sugar beet varieties and two fungicide strategies in a two-factorial block design with four blocks or repetitions. The two fungicide strategies are: control with fungicide, and inoculated without fungicide. The registration identifiers of the five sugar beet varieties Beta vulgaris L. ssp. vulgaris (A–E) are: 3012, 2444, 3290, 3316 and 3706 respectively. Selected varieties belong to the national variety list of the German Federal Plant Variety Office (Bundessortenamt) [23].
Disease assessment: We carry out visual scoring of DS of CLS as ground truth simultaneously to the UAV flights, and quantify symptoms as an average value of the plot by assessing middle leaves in percentage estimating a representative infected from total leaf area [6]. The assessment is conducted at leaf level. In total, we randomly sample 100 leaves (25 leaves/plot) per treatment and inspect them for CLS symptoms.
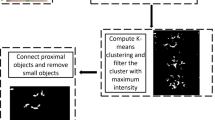
Prediction on large images: Orthomosaic images from UAV-imagery outrun current GPUs capacity for prediction. To extend the model application to a large image from a complete mapped field, we use a sliding windows procedure (Fig. 2). For this purpose and as proposed by Machefer et al. [24], we crop the RGB composite orthomosaic image (I) in patches with an overlap (15 cm or 60 px, o) between neighbours. We use the segmentation model to predict leave instances sequentially on each patch; and eliminate instances with 100% cover in the overlap region. After selection, we reconstruct the remaining leaves in a new orthomosaic image (\(I_{leaf}\)). Algorithm 1 shows the procedure.
Instance segmentation for leaf parameter extraction: The availability of \(I_{leaf}\) increases the potential for extraction of plant phenotyping parameters. In this study, we propose the integration of this output to a pipeline for pixel-wise classification and extraction of disease-relevant parameters as specified by Barreto et al. [11]. The new pipeline starts with radiometric calibration and photogrammetric processing of raw images resulting in the multispectral orthomosaic and the digital surface model (DSM). Then, we create a RGB composite orthomosaic image as specified in Sect. 2.1 (Fig. 3a). In the next step, prediction of large image takes place to deliver the \(I_{leaf}\) output (Fig. 3b). We calculate image features of multispectral orthomosaic and DSM and store them for later feeding them to two multiclass classifiers, a partial least squares discriminant analysis (PLS-DA), and a support vector machine radial (SVMR). At this level each pixel (Z) is assigned one of four classes, “other", “soil", “diseased", “healthy". This results in a binary array of four layers or images: \(O_{i,j}\), \(S_{i,j}\), \(D_{i,j}\), and \(H_{i,j}\), assigned to the four classes respectively. The overlay of any of those outputs with an instance L (\(L \in I_{leaf}\)) returns four outputs with the delimited instance region: \(O_{L}\), \(S_{L}\), \(D_{L}\), and \(H_{L}\) (Fig. 3c). The binary image \(D_{L}\) is relevant for disease quantification; therefore, clusters (c) can be extracted from \(D_{L}\) (Fig. 3c) by labeling eight connected cluster pixels [25].
Considering the image area (\(A_{L}\)) and image slope (\(\zeta _{L}\)) as calculated features from DSM within an instance L [11, 26], we calculate the leaf-based parameters with the formulae in Table 4. These parameters are: leaf area (\(A_{l}\)), leaf slope (\(\zeta _l\)), diseased leaf area (\(A_{D}\)), healthy leaf area (\(A_{H}\)), disease severity (cover based, \(ds_{l}\)), disease severity (area based, \(DS_{l}\)), number of clusters (c), and mean cluster area (\(A_{\overline{c}}\)).
Automatic scoring at plot level: Some parameters can not be directly determined from one instance, but they require a sample of instances within a field area. In field trials, this sampling takes place within a plot. A representative sampling for monitoring diseases normally consists of taking 100 leaves for scoring per plot [6]. In the same way, we can express the parameters mentioned in Table 4 as an average within the field area. Moreover, we can calculate the relevant parameter DI in a plot-based manner. To determine DI, it is necessary to define an affected or diseased unit as mentioned by [11, 27, 28]. In the past, we defined an instance as diseased if at least one lesion was present or if the pixel summation in D was greater or equal than one cluster (\(c \ge 1\)). We can also define a diseased unit with another parameter such as \(ds_{l}\). Similarly, it is possible to establish threshold values to order diseased leaves (Fig. 6a).
3.2 UAV Parameter for Disease Quantification
Leaf segmentation for disease severity: We compare the disease quantification parameter, DS from UAV-based and expert-based sources in Fig. 4. Figure 4a and b show a parabolic behavior of the UAV-based DS for models with the lowest and the highest AP (strategy I and V). This behaviour presents a maximum value of UAV-based DS at 40% of the ground truth data, and seems to be a limit for the automatic calculation of leaf-based DS. By evaluating the proportion of variance in linear regression (\(r^{2}\)) between both DSs, we observe slightly better results in the linear regression model of the UAV-based data from strategy V compared to strategy I. Moreover, there are no clear differences between models with CS of 0.50 or 0.75.
We attribute this parabolic relationship of UAV-based DS to the image context and the combined effect from nadir UAV perspective and the new leaf growth emerging of a typical severe epidemics of CLS [29]. The nadir perspective of UAV record mainly above-located leaves which are under severe epidemics and covered by healthy new leaves. This entails special advantage of visual scoring because from a side perspective heavy diseased and small leaves are quantified with high DS values [30], while low DS values are delivered from UAV-based scores by quantifying healthy new leaves.
To mitigate the effect of image context by analyzing less disease development stages, in Fig. 4c and d were deleted all data with high expert-based values (DS higher than 40%). In general, values of \(r^{2}\) increase significantly after data deletion. Here, the model with the highest AP (strategy V) shows a clear advantage against model of strategy I. In addition, the model with CS of 0.75 performs slightly better than the model with CS of 0.50. A possible solution against this image context challenge is to introduce a new class of leaf instances for leaves with damage higher than 40% in order to give priority to the detection of this morphological different leaves.
Epidemiological development of disease quantification parameters: In this section we show one of the principal applications of leaf segmentation for parameter extraction and plant phenotyping of variety trials for resistance quantification. As described in Sect. 3.1, we work with the experimental design with five varieties and two fungicide strategies. In Fig. 5, we show the development of each genotype during the complete period for disease monitoring with average values of \(DS_{l}\), \(A_{l}\), and c. We compare the development of \(DS_{l}\) in Fig. 5a and b for the control with fungicide and inoculated without fungicide variants. Here we see the effect of disease pressure by applications of fungicide, where the control with fungicide variant keeps all varieties healthy until the beginning of September (Fig. 5a). We further observe resistant characteristics of a genotype with high disease pressure. In Fig. 5b, variety A shows the most susceptible characteristics, and variety E presents apparently the highest tolerance against CLS. The leaf area is also affected by the genotype and disease pressure (Fig. 5c and d). In Fig. 5c we see that variety C presents the smallest \(A_{l}\) until mid August, while varieties D and E show the biggest \(A_{l}\) during the complete monitoring period. \(A_{l}\) is further affected by disease pressure. UAV-monitoring is a new way to describe resistance (Fig. 5d), where an accumulative value of leaf area highlights the resistance of a genotype. Figure 5e and f show the development of c, where we observe similar properties for disease quantification as with \(DS_{l}\). The shape for instance should influence the relevance of c for disease quantification. In our past contribution [11], we calculate c from a circle shape instance and do not find relevance for disease quantification and variety differentiation for resistance. However, using leaf form instances from our instance segmentation model, we are able to contrast variety quantitative resistance.
UAV-parameters for plant protection measure: In Germany, thresholds of DI are the principal indicators for plant protection measures, avoiding losses in sugar beet cultivation [6]. On the field and for the case of CLS, leaves are ordered as “diseased" when at least one CLS spot is present on a sampled middle leaf. To implement an automatic scoring approach, an accurate and robust detection of single spots is compulsory. However, current pixel-wise UAV-based image processing approaches are far from 100% level of precision and recall [9, 11]. This disadvantage makes the automatic definition of a diseased unit a challenge due to the risk of false positives if the criteria “\(c \ge 1\)" is considered, because of the wrong classification of healthy regions as diseased increases. However, this criterium is easy to adapt to different disease quantification parameters in image-processing approaches (e.g. by using c or \(ds_{l}\)) and diverse threshold values (Fig. 6a). We emulate possible time-series developments of DI with four different threshold values that defines a diseased unit ( “diseased" \(if\{c \mid ds_{l} \ge 1, 5, 10, 25\}\) number of clusters or %). In Fig. 6b we observe that DI based on c parameter has a slightly advantage in comparison with DIs based on \(ds_{l}\) criteria, due to the early exponential phase in DI curve. Nevertheless, DI with the most sensible definition criteria of “diseased” (“diseased” if \(\{c \ge 1\}\)) shows the disadvantage of the inaccurate pixel-wise approach, delivering high values of DI at the begin of July when the pathogen was just inoculated, with false positive detections. This inaccuracy can lead to a wrong decision for disease control. The alternative to this problem is to fix a higher threshold value to define a diseased unit avoiding possible emergence of false positives as observed in criteria \(\{c \ge 5, 10, 25\}\). However, this decision shifts the exponential phase of DI to a later point in time, making this adaptation not suitable for practical use.
In Fig. 6c, we visualize the development of expert-based and UAV-based parameters. We observe that DS from expert-based and our leaf segmentation UAV-based data stay close together. This demonstrates that multispectral UAV-imaging for an automatic DS assessment under field conditions is feasible. However, in the case of DI, the expert ratings show an early exponential phase in the curve in July, while UAV-based scoring with a criteria “\(c \ge 5\)” presents an exponential phase two months later than the experts. The performance of UAV-based DI parameter can be improved with a higher precision and recall for detecting diseased regions. However, we suspect there is a technical gap in the case of CLS for multispectral VISNIR UAV-imaging comparison to expert evaluation. We believe that this is due to restricted view of nadir perspective of UAV-based images in comparison to the possibility of the expert to sample middle and old sugar beet leaves individually, where first symptoms are observed [31].
4 General Discussions, Further Work and Conclusion
In this work we give an overview of the complexity of modeling a leaf segmentation approach for UAV-images under field condition. For practical use, automatic leaf segmentation should provide robustness. This includes good performance under diverse illumination conditions, for a high number of genotypes, and considering the epidemiological development of infested plants. This requires a high number of annotated images for modeling one plant–pathogen interaction.
Mask R-CNN models with a good AP performance deliver more accurate disease quantification parameters according to our results in comparison with models with a low AP. Leaf-based phenotyping parameters are relevant tools to describe epidemiological development of sugar beet genotypes under diverse disease pressure. Accumulated \(A_{l}\)Ó over the complete monitoring season should differentiate genotypes for resistance against leaf diseases.
Instance form, whether circle or leaf, should influence the relevance of respective parameters to differentiate genotypes. To graph this statement, we evaluate the case for c parameter from our past contribution [11], where circle-form c showed no significance for the genotype factor, while a leaf-form c presents a potential for resistance differentiation (Fig. 5c).
One of the principal limits for monitoring and scoring leaf diseases in field experiments is the necessity of a high number of trained personal. An automatic UAV-based scoring offers the chance to eliminate human error proper from individual scoring increasing the efficiency of collecting data.
The potential application of multispectral UAV-imaging and leaf-based parameters for disease management has to be analyzed in future studies. Although we mention the possible technical gap for UAV-based DI, we have to consider the potential of dividing a mapped field in mini plots for calculating DI, to make an individual and geo-referenced decision in large mapped fields. Reduction of fungicide use is an important step towards sustainable sugar beet production. This reduction is feasible through site-specific applications guided by geo-referenced and accurate UAV-based DI’s as indicators for decision-making.
In conclusion, our experiments show basic image augmentation techniques to be more efficient than the computationally expensive copy-paste augmentation approach for training the leaf segmentation Mask R-CNN on UAV-images in our resource constraint setting. We demonstrate that a leaf-segmentation Mask R-CNN model can be integrated in a pipeline to extract leaf-based parameters of monitored fields. Advantages and limitations from calculated parameters are identified and critically discussed. Moreover, geo-referenced decision making and site-specific application needs to be addressed by future research. Although we evaluate only the case of CLS disease, the present pipeline for leaf-based parameter extraction can be transferred to agricultural practice and can support decision making in plant breeding for resistance and integrated disease management. Overall, we demonstrate that UAV-based monitoring of sugar beet fields followed by proper post-processing, can output reliable information that increases efficiency by replacing the very laborious work of visual scoring.
Prediction of disease quantification parameters by application of leaf segmentation. a RGB composite of diseased plants, b segmentation of leaves, and c multiclass pixel-classification and parameter extraction of leaf instances as healthy region (\(H_{L}\)), diseased region (\(D_{L}\)), and number of clusters within a leaf (c)
Relationship of unmanned aerial vehicle (UAV)- and expert-based scores for confidence scores (CS) of 0.50 and 0.75 in two monitoring dates of experimental test field. Comparison of all ground-truth values for model in data augmentation a strategy I and b strategy V. Comparison for ground truths lower than 40 % in c strategy I and d strategy V. Monitoring dates: 17th and 31st August 2021
Development within fungicide strategies and five sugar beet varieties of leaf-based disease quantification parameters: disease severity (a, b), leaf area (c, d), and number of clusters per leaf (e, f). A total of 100 leaves were used for each treatment level. Curves were smoothed using locally estimated scatterplot method
Decision making based on disease incidence (DI). a Criteria for defining a diseased unit based by thresholding c or \(ds_{l}\) parameter. b Development of DI curve based on diverse threshold magnitudes individually for c and \(ds_{l}\) parameters. c Development of UAV-based and expert-based parameters. A total of 100 leaves were used for the treatment level in time-series (variety B and control with fungicide). Curves were smoothed using locally estimated scatterplot method
Data availability
The multispectral UAV image data, along with the visual scoring data, were acquired by the Institute of Sugar Beet Research (IfZ) as part of the Coordination Beet Research International (COBRI) project titled ‘Sensing of Plant Diseases by Hyperspectral Imaging and UAVs’. Requests for data may be submitted to IfZ through the corresponding author.
References
Aasen H, Honkavaara E, Lucieer A, Zarco-Tejada PJ (2018) Quantitative remote sensing at ultra-high resolution with UAV spectroscopy: a review of sensor technology, measurement procedures, and data correction workflows. Remote Sens. https://doi.org/10.3390/rs10071091
Maes WH, Steppe K (2019) Perspectives for remote sensing with unmanned aerial vehicles in precision agriculture. Trends Plant Sci 24(2):152–164. https://doi.org/10.1016/j.tplants.2018.11.007
Vereijssen J, Schneider JHM, Termorshuizen AJ, Jeger MJ (2003) Comparison of two disease assessment methods for assessing Cercospora leaf spot in sugar beet. Crop Prot 22(1):201–209. https://doi.org/10.1016/S0261-2194(02)00146-1
Madden LV, Hughes G, van den Bosch F (2007) Temporal analysis I: quantifying and comparing epidemics. In: The Study of Plant Disease Epidemics, pp 63–116
Bock C, Barbedo JGA, Del Ponte EM, Bohnenkamp D, Mahlein A-K (2020) From visual estimates to fully automated sensor-based measurements of plant disease severity: status and challenges for improving accuracy. Phytopathol Res 2(1):9. https://doi.org/10.1186/s42483-020-00049-8
Wolf PFJ, Verreet JA (2002) An integrated pest management system in Germany for the control of fungal leaf diseases in sugar beet: the IPM sugar beet model. Plant Dis 86(4):336–344. https://doi.org/10.1094/pdis.2002.86.4.336
Reynolds D, Baret F, Welcker C, Bostrom A, Ball J, Cellini F, Lorence A, Chawade A, Khafif M, Noshita K, Mueller-Linow M, Zhou J, Tardieu F (2019) What is cost-efficient phenotyping? optimizing costs for different scenarios. Plant Science 282:14–22. https://doi.org/10.1016/j.plantsci.2018.06.015. The 4th International Plant Phenotyping Symposium
Jay S, Comar A, Benicio R, Beauvois J, Dutartre D, Daubige G, Li W, Labrosse J, Thomas S, Henry N, Weiss M, Baret F (2020) Scoring Cercospora leaf spot on sugar beet: Comparison of UGV and UAV phenotyping systems. Plant Phenomics 2020, 1–18 . https://doi.org/10.34133/2020/9452123
Görlich F, Marks E, Mahlein A-K, König K, Lottes P, Stachniss C (2021) UAV-based classification of Cercospora leaf spot using RGB images. Drones 5(2) . https://doi.org/10.3390/drones5020034
Yamati FRI, Barreto A, Günder M, Bauckhage C, Mahlein A-K (2022) Sensing the occurrence and dynamics of cercospora leaf spot disease using UAV-supported image data and deep learning. Sugar Industry, 79–86 . https://doi.org/10.36961/si28345
Barreto A, Ispizua Yamati FR, Varrelmann M, Paulus S, Mahlein A-K (2022) Disease incidence and severity of Cercospora leaf spot in sugar beet assessed by multispectral unmanned aerial images and machine learning. Plant Disease
Günder M, Ispizua Yamati FR, Kierdorf J, Roscher R, Mahlein A-K, Bauckhage C (2022) Agricultural plant cataloging and establishment of a data framework from UAV-based crop images by computer vision. GigaScience 11 https://academic.oup.com/gigascience/article-pdf/doi/10.1093/gigascience/giac054/44118901/giac054.pdf. https://doi.org/10.1093/gigascience/giac054. giac054
He K, Gkioxari G, Dollár P, Girshick R (2018) Mask R-CNN
Danielczuk M, Matl M, Gupta S, Li A, Lee A, Mahler J, Goldberg K (2019) Segmenting Unknown 3D Objects from Real Depth Images using Mask R-CNN Trained on Synthetic Data. In: 2019 International Conference on Robotics and Automation (ICRA), pp. 7283–7290 (2019). https://doi.org/10.1109/ICRA.2019.8793744
Kuznichov D, Zvirin A, Honen Y, Kimmel R (2019) Data Augmentation for Leaf Segmentation and Counting Tasks in Rosette Plants. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops
Barreto A, Lottes P, Ispizua Yamati FR, Baumgarten S, Wolf NA, Stachniss C, Mahlein A-K, Paulus S (2021) Automatic UAV-based counting of seedlings in sugar-beet field and extension to maize and strawberry. Comput Electron Agric 191:106493. https://doi.org/10.1016/j.compag.2021.106493
Abdulla W (2017) Mask R-CNN for object detection and instance segmentation on Keras and TensorFlow. Github
Leng J, Ren Y, Jiang W, Sun X, Wang Y (2021) Realize your surroundings: Exploiting context information for small object detection. Neurocomputing 433:287–299. https://doi.org/10.1016/j.neucom.2020.12.093
Chen C, Liu M-Y, Tuzel O, Xiao J (2017) R-cnn for small object detection. In: Lai, S.-H., Lepetit, V., Nishino, K., Sato, Y. (eds.) pp. 214–230. Springer, ???
Tong K, Wu Y, Zhou F (2020) Recent advances in small object detection based on deep learning: A review. Image Vis Comput 97:103910. https://doi.org/10.1016/j.imavis.2020.103910
Weyler J, Magistri F, Seitz P, Behley J, Stachniss, C (2022) In-field phenotyping based on crop leaf and plant instance segmentation, 2725–2734
Liu L, Ouyang W, Wang X, Fieguth P, Chen J, Liu X, Pietikäinen M (2020) Deep learning for generic object detection: A survey. Int J Comput Vision 128:261–318. https://doi.org/10.1007/s11263-019-01247-4
Bundessortenamt (2022): Getreide, Mais, Öl- und Faserpflanzen. In: Beschreibende Sortenliste, p. 369
Machefer M, Lemarchand F, Bonnefond V, Hitchins A, Sidiropoulos P (2020) Mask R-CNN refitting strategy for plant counting and sizing in uav imagery. Remote Sensing 12(18):23. https://doi.org/10.3390/RS12183015
He L, Ren X, Gao Q, Zhao X, Yao B, Chao Y (2017) The connected-component labeling problem: A review of state-of-the-art algorithms. Pattern Recogn 70:25–43. https://doi.org/10.1016/j.patcog.2017.04.018
Corripio JG (2003) Vectorial algebra algorithms for calculating terrain parameters from DEMs and solar radiation modelling in mountainous terrain. Int J Geogr Inf Sci 17(1):1–23. https://doi.org/10.1080/713811744
Nutter F Jr, Teng P, Shokes FM (1991) Disease assessment terms and concepts. Plant Dis 75:1187–1188. https://doi.org/10.1094/PD-75-1187
Bock CH, Pethybridge SJ, Barbedo JGA, Esker PD, Mahlein A-K, Ponte EMD (2021) A phytopathometry glossary for the twenty-first century : towards consistency and precision in intra- and inter-disciplinary dialogues. Tropical Plant Pathology. https://doi.org/10.1007/s40858-021-00454-0
Weiland J, Koch G. (2004) Sugarbeet leaf spot disease (Cercospora beticola Sacc.). Molecular Plant Pathology 5(3), 157–166 . https://doi.org/10.1111/j.1364-3703.2004.00218.x
Woo HR, Kim HJ, Lim PO, Nam HG (2019) Leaf senescence: Systems and dynamics aspects. Annu Rev Plant Biol 70:347–376. https://doi.org/10.1146/annurev-arplant-050718-095859
Rangel LI, Spanner RE, Ebert MK, Pethybridge SJ, Stukenbrock EH, de Jonge R, Secor GA, Bolton MD (2020) Cercospora beticola: The intoxicating lifestyle of the leaf spot pathogen of sugar beet. Mol Plant Pathol 21(8):1020–1041. https://doi.org/10.1111/mpp.12962
Acknowledgements
This research was partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy - EXC 2070-390732324 and supported by funds of the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the Parliament of the Federal Republic of Germany. The Federal Office for Agriculture and Food (BLE) provides coordinating support for artificial intelligence (AI) in agriculture as funding organisation, grant number FKZ 28DE104A18. This work was also financially supported within Coordination Beet Research International (COBRI).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Appendix A Supplementary information
Appendix A Supplementary information
- UAV:
-
Unmanned aerial vehicle
- CLS:
-
Cercospora leaf spot
- DSM:
-
Digital surface model
- DI:
-
Disease incidence
- DS:
-
Disease severity
- AP:
-
Average precision
- ROI:
-
Region of interest
- ML:
-
Machine learning
- IoU:
-
Intersection over union
- CS:
-
Confidence score
- GSD:
-
Ground sample distance
- A:
-
Surface area
- \(\zeta _{L}\):
-
Image slope or angle between surface and normal to horizontal within a leaf instance
- \(\zeta _l\):
-
Average slope or angle between surface and normal to horizontal within a leaf instance
- PLS-DA:
-
Partial least squares discriminant analysis
- SVMR:
-
Support vector machine radial
- L:
-
Individual leaf instance
- \(A_{L}\):
-
Image surface area within leaf instance
- \(A_{l}\):
-
Surface area within leaf instance
- \(A_{\overline{c}}\):
-
Average cluster area within leaf instance
- \(A_{H}\):
-
Surface area of healthy foliage within leaf instance
- \(A_{D}\):
-
Surface area of diseased foliage within leaf instance
- \(ds_{l}\):
-
Cover based disease severity within leaf instance
- \(DS_{l}\):
-
Area based disease severity within leaf instance
- W:
-
Trained weights for leaf segmentation Mask R-CNN model
- c:
-
Number of clusters
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Barreto, A., Reifenrath, L., Vogg, R. et al. Data Augmentation for Mask-Based Leaf Segmentation of UAV-Images as a Basis to Extract Leaf-Based Phenotyping Parameters. Künstl Intell 37, 143–156 (2023). https://doi.org/10.1007/s13218-023-00815-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13218-023-00815-8