Abstract
Exploiting mutual explanations for interactive learning is presented as part of an interdisciplinary research project on transparent machine learning for medical decision support. Focus of the project is to combine deep learning black box approaches with interpretable machine learning for classification of different types of medical images to combine the predictive accuracy of deep learning and the transparency and comprehensibility of interpretable models. Specifically, we present an extension of the Inductive Logic Programming system Aleph to allow for interactive learning. Medical experts can ask for verbal explanations. They can correct classification decisions and in addition can also correct the explanations. Thereby, expert knowledge can be taken into account in form of constraints for model adaption.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Medical decision making is one of the most relevant real world domains where intelligent support is necessary to help human experts master the ever growing complexity. Since medicine is a highly sensitive domain where errors can lead to fatal errors, transparency and comprehensibilty are legal as well as and ethical requirements [24]. Therefore, the usage of standard approaches of machine learning, such as (deep) neural networks, is not recommendable because the learned models are blackbox [1]. That is, the user has only access to the input information (for instance a medical image) and the resulting classifier decision as output. The reasoning underlying this decision remains intransparent. Another challenge when applying machine learning in medicine and in many other real world domains is that the amount and quality of data often cannot meet the demands of highly data intensive machine learning approaches: Classes are often strongly imbalanced and for many specific manifestations of clinical diagnoses data are sparse. Apart from routine diagnoses, in many cases there is no ground truth available. Diagnostic gold standard tests often have limitations in reliability as well as validity.
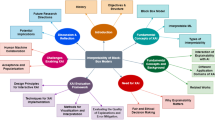
The ultima ratio to overcome this data engineering bottleneck is to involve human who have the expertise to evaluate quality of data as well as validity of the output of learned models. In consequence, incremental and interactive approaches are promising options for making use of machine learning in medical diagnostics [13]. Starting with an initial model, new cases can be incorporated as they occur in practice, and system decisions based on erroneous labeling can be corrected in the context of a current application. While class correction is standard in interactive learning [9, 32], we propose to exploit explanations to constrain model adaptation. That is, we do not consider explanations as a one-way street from the system to the user but provide a method for mutual explanations as a necessary step towards a balanced human-AI partnership (see Fig. 1).
In the following, we present the research project Transparent Medical Expert Companion in which we aim at developing an approach for such a balanced human-AI partnership by making machine learning based decisions in medicine transparent, comprehensible, and correctable. The main outcome of the project will be a framework for an explanation interface which is based on mutual explanations. This framework will be instantiated for two application domains—colon cancer diagnosis from tissue scans and pain assessment from video sequences [29]. We introduce the colon cancer use case in the next section. Afterwards, we introduce Inductive Logic Programming (ILP) as powerful approach of interpretable machine learning which naturally allows to combine reasoning and learning. In the next section we present the different approaches of explanation generation we investigate for comprehensible decision making—visual, verbal, and contrastive explanations. The focus of this paper is to give an overview of the constituents of our framework. In addition, we present how mutual explanations can be realised by extending the ILP system Aleph [30]. This extension allows the medical expert to correct explanations to constrain model adaption.
2 Image Based Medical Data with Spatial Relations
Medical diagnosis in many domains relies strongly on image data such as radiographic, ultrasonic or microscopic images. Furthermore, analyses of bio-medical signals such as cardiograms as well as high-level behavioral data from videos rely on visual inspection [23]. To analyze image-based data, human experts often take into account spatial information. In colon cancer diagnosis, medical experts analyze the tissue composition and the depth of invasion of tumors. For instance, if tumor tissue already touches fat, the tumor class is more critical compared to a situation where the tumor is included in fascial tissue [35]. In consequence, machine learning approaches should be able to reveal which relationships among tissues have lead to a certain classification of a microscopic image. These relationships also must be communicated to the medical decision maker in a comprehensible way. While the position of the tumor can be marked in an image, the relationship can better be expressed in natural language [25].
In Fig. 2 we present our mutual explanation interface. In the upper part, a selection of tissue scans is presented which have been classified—for instance by convolutional neural network classifier (CNN). Four scans have been classified as tumor class pt3 and the ILP learner induced a model characterising these scans in contrast to two scans classified as healthy. An expert pathologist inspects the learned rules given in the bottom of the interface and discovers that one of the rules contains an erroneous touches relation. He or she marks the erroneous part and can define the constraint that this part should excluded in future models (see bottom middle of the interface). The model is updated and as result to scans previously classified as pt3 are now moved to the negative examples (see top right of the interface). The expert can inspect these scans and either change their label or modify the rules again.
3 Interpretable Machine Learning with ILP
Current deep neural network approaches which allow end-to-end learning from raw data without the need of feature extraction have shown to result in models with very high predictive accuracy. The most impressive results have been gained with CNNs for image classification [14, 16]. On the other hand—in contrast to these black box approaches—there are interpretable (white box) approaches where learned models are represented in a symbolic, human readable, explicit form [8]. Typical white box approaches are variations of decision tree learning [11, 15]. Decision trees and other models represented as rule allow for straight-forward rewriting of reasoning traces into natural language explanations. Such a procedure has already been proposed in early AI in the context of expert system research for the system MYCIN which supported diagnosis of bacterial infections [4].
A more general representation format than decision rules is offered by first order logic. Here, rules can be defined over variables and it is possible to express arbitrary relations. Inductive Logic Programming (ILP) allows to learn models composed of such logical rules [21]. ILP allows to combine reasoning and learning in a natural way. Background theories can be exploited during learning and learned rules can be combined with optionally predefined rules for inference. It has been shown that rules learned with ILP can support human decision making in complex domains [22]. Transforming such rules into verbal explanations can be done with similar methods as have been introduced in the context of expert systems [28].
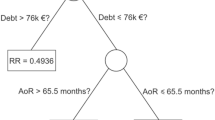
A simple example is given in Fig. 3. Spatial relations can be defined by topological models defined over points, lines and polygonal areas [3]. For example, it can be defined what it means that two areas are touching each other, are disjoint, or that one area includes another one. Such relations are meaningful to medical experts which are used to characterize tumor classes by the size and location of the tumor in relation to other types of tissue [35]. In contrast to classical machine learning approaches such as decision trees, examples are not given as feature vectors but as structural representations. Every feature vector representation can be transformed into such a structural representation without loss of information but not the other way round. In the context of classification of mutagenicity, it has been shown that the richer structural representations result in significantly higher accuracy in contrast to simple features [31].
4 Visual, Verbal, and Contrastive Explanations
Explanations in human–human interaction have the function to make something clear by giving a detailed description, a reason, or justification [17, 18]. In the context of explaining decisions of black box classifiers, there is a strong focus on visual explanations where areas relevant for a decision are highlighted [26, 33]. Visual highlighting can be helpful for machine learning experts to recognize problems such as overfitting. Furthermore, it allows for fast communication by directing attention. However, for comprehending complex medical images, highlighting alone is not expressive enough [27]: The only visible relation between two areas corresponds to a conjunction (there is fat and tumor), spatial relations cannot be expressed (tumor tissue is touching fat tissue). Furthermore, —although there is work on visualizing negative evidence [26]—the absence of something cannot in general be captured visually (There is no tumor tissue). Finally, often it is not sufficient to explain what part of an image is relevant for a decision. For example, nearly all facial expressions involve areas around the eyes, the mouth, and the nose. To understand why a classifier identified a specific emotion, it is relevant to have information not about a feature (eye) but about the feature value (lid is tightened indicating pain vs. upper lid is raised indicating surprise).
Sometimes, it is difficult to grasp the expression of a feature. In this case, contrastive examples are especially helpful. For instance, in a psychological experiment, Gentner and Markman [12] could show that explaining what a lightbulb is, is easier when contrasting it with a candle compared to contrasting it with a cat. Recently, contrastive explanations have also been proposed for black box image classification [7]. This is closely related to learning structural descriptions from near misses (most similar instance not belonging to the target class) which as been shown to make learning more efficient [34].
To support joint decision making in medical diagnosis, we propose that it is recommendable to offer a variety of explanation styles. Currently we focus on combining visual and verbal explanations for the current instance as well as a near miss example. We exploit different image segmentation methods to relate parts of the image with relations and attributes captured by the ILP learned rules with a focus on different superpixel approaches [25]. How pain classification can be explained by constrasting a painful expression with disgust is introduced in Schmid [27].
5 Exploiting Mutual Explanations for Learning
Dialogue systems have been originally proposed for knowledge-based systems [2] and could be used to realize joint decision making. Human interaction can serve as a model. Here, one human might ask another to explain his or her decision. For example, one medical expert might ask another for the reasons behind a pT3 diagnosis (see Fig. 2). The given explanation can be accepted by the other person or not. In case of rejection, it can be indicated which parts of the given explanations are not acceptable. Together the discussion partners can find an alternative explanation. We propose that this dialogue-based, incremental process should be captured by explainable AI methods. Such mutual explanations are cooperative, interactive, and incremental acts of information exchange between humans and machines with the goal to improve the joint performance of the involved partners in classification problems. We propose that the process of explanation refers to providing arguments [20] that make simple and complex relations, which apply to the domain of interest, explicit. It further refers to integrating corrective explanations into existing internal models in order to adapt these [10].
A model of such a mutual explanation system is given in Fig. 1: Starting with an initial ILP model, a new example e is classified. The class decision is presented to the human who can confirm it or ask for an explanation. The explanation can be accepted or not. In case of rejection, the human can correct the explanation. The correction together with the new class label are integrated to adapt the model. While it is possible that a correct classification can be associated with a wrong or insufficient explanation, we focus on correcting explanations associated with erroneous class decisions. The proposed approach is applicable to both cases.
A wrong class decision can be either a miss or a false alarm. In machine learning, this can be attributed to overly specific or overly general model [6, 19]. In ILP, a learned model \(\mathcal {M}\) for a single target predicate consists of first order rules R of the following form:
where the head of the rule is an atom and the body is a conjunction of literals. Rules are defined over variables. An instance is a conjunction K of ground literals. An instance is classified as member of the target concept, if there exists a substitution \(\theta \) of variables in a rule \(R \in \mathcal {M}\) such that \(K \subseteq body (R)\theta \).
As described in De Raedt [5] theta-subsumption can be considered to be a constraint satisfaction problem. Hence, human corrections of explanations can involve adding or deleting literals or restricting or enlarging the scope of arguments.
More formally, given a conjunction of literals (\(l_{1}\)\(\wedge \) ...\(\wedge \)\(l_{n}\)) from \( body (R)\) and a conjunction of boolean constraints (\(c_{1}\)\(\wedge \) ...\(\wedge \)\(c_{n}\)), a substitution \(\theta \in \{\top , \bot \}\) needs to be found for every literal, such that the resulting boolean formula \(L_{i} \wedge C_{n}\) evaluates to \(\top \), if \(L_{i}\) is a valid clause to be added to the theory, \(\bot \) otherwise. Each \( body (R)\) that satisfies \(C_{n}\) can be added to the theory, if it has the best score with respect to Aleph’s evaluation setting. Given a literal’s set of arguments (\(a_{1} \wedge \ldots \wedge a_{n}\)), a domain \(D(a_{i})\) for every \(a_{i}\) and a conjunction of numerical or set constraints, a substitution \(\theta \) needs to be found, such that \(D(a_{i})\)\(\ge _{g}\)\(D(a_{i} \theta )\) for overly general clauses and \(D(a_{i} \theta )\)\(\ge _{g}\)\(D(a_{i})\) for overly specific clauses. For overly specific clauses, \(\theta \) substitutes constants with variables. For overly general clauses, \(\theta \) substitutes variables with constants or different variables, which are already present in the set of arguments.
An overly general model can result in a false alarm, erroneously classifying an instance as member of the target concept—such as tumor class pT3 or pain. A human expert might introduce an additional literal or restrict the scope of a predicate. For instance, a rule

is too general and can be restricted by introducing

A rule might specify the size of a tumor in millimeters

which also can be restricted in case of over-generality of a rule by requiring the value to be larger than 5.
An overly specific model can result in misses. For instance, the rule

excludes tumors from class pT3 where tumor and muscle tissue are not disjoint. Likewise, specific values, for instance, the size of a tumor, might be too restrictive. Decreasing the minimum size of a tumor makes a rule more general.
In Aleph, user-defined constraints can be applied to guide the generation of alternative clauses [30]. To make the interaction with our system easier for medical experts, they can mark parts of an explanation, which are then transformed into proper constraint syntax. For example, if we require a clause to contain some predicate p with arguments X and a, where X is a variable and a is a constant, a typical constraint is represented as follows:

The head of a constraint is set to false. This way, all clauses evaluate to false, where the goals in the body of the constraint are satisfied. The constraint above expresses that a body containing p must occur in a clause. The set of user-defined constraints and the current clause are combined into a boolean formula for SAT solving as well as unification is performed. Aleph then generates new rule candidates, considering only the ones which satisfy the constraints for theory construction.
We conducted a first experiment to evaluate our mutual explanation approach (see Finzel [10] for details). We generated a small artificial data set for the colon cancer domain and introduced erroneous class labels which resulted in false positives. We iteratively applied boolean constraints for corrections of erroneous explanations at the clause level. At the literal level we applied set and numerical constraints. All constraints were generated from user feedback via an explanation interface as shown in Fig. 2.
Applying the constraints led to a specialization of the induced theory and thus the exclusion of false positives. Results further indicate that introducing constraints can help to decrease the necessary number of corrections. However, corrections can result in higher computational effort during search. This preliminary evaluation can be seen as a first proof of concept. We currently are conducting an evaluation with a larger data set where we assess the reduction of errors and computational time in a systematic way.
6 Conclusions
We presented a framework for making use of mutual explanations for joint decision making in medicine. Inductive Logic Programming was introduced as an expressive approach of interpretable machine learning which allows to make use of domain knowledge in learning and inference. We discussed the merit of different types of explanations and argued for combining visual and verbal explanations to allow for conveying complex relational information as well as absence of features and presence of specific feature values.
We argued that explanations should be provided by the system to the human but also the other way round and gave a sketch how explanations can be applied as constraints for the ILP system Aleph. Based on promising first results, we plan to evaluate this approach more extensively for several medical image domains.
References
Adadi A, Berrada M (2018) Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access 6:52138–52160
Bickmore T, Giorgino T (2006) Health dialog systems for patients and consumers. J Biomed Inf 39(5):556–571
Chen J, Li C, Li Z, Gold C (2001) A voronoi-based 9-intersection model for spatial relations. Int J Geogr Inf Sci 15(3):201–220
Clancey WJ (1983) The epistemology of a rule-based expert system’a framework for explanation. Artif Intell 20(3):215–251
De Raedt L (2008) Computational aspects of logical and relational learning. In: De Raedt L (ed) Logical and relational learning. Springer, Heidelberg, pp 325–343
De Raedt L, Kramer S (2001) The levelwise version space algorithm and its application to molecular fragment finding. In: Proceedings of the 17th international joint conference on artificial intelligence, pp 853–862. Morgan Kaufman
Dhurandhar A, Chen P-Y, Luss R, Tu C-C, Ting P, Shanmugam K, Das P (2018) Explanations based on the missing: towards contrastive explanations with pertinent negatives. In: Advances in neural information processing systems, pp 592–603
Doshi-Velez F, Kim B (2017) Towards a rigorous science of interpretable machine learning. arXiv:1702.08608
Fails JA, Olsen Jr DR (2003) Interactive machine learning. In: Proceedings of the 8th international conference on intelligent user interfaces, ACM, pp 39–45
Finzel B (2019) Explanation-guided constraint generation for an inverse entailment algorithm. Master’s thesis, University of Bamberg
Fürnkranz J, Kliegr T (2015) A brief overview of rule learning. In: International symposium on rules and rule markup languages for the semantic web, pp 54–69, Springer, New York
Gentner D, Markman AB (1994) Structural alignment in comparison: no difference without similarity. Psychol Sci 5(3):152–158
Holzinger A (2014) Trends in interactive knowledge discovery for personalized medicine: cognitive science meets machine learning. IEEE Intell Inf Bull 15(1):6–14
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
Lakkaraju H, Bach SH, Leskovec J (2016) Interpretable decision sets: a joint framework for description and prediction. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, ACM, pp 1675–1684
Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M (2014) Medical image classification with convolutional neural network. In: 2014 13th International conference on control automation robotics and vision, IEEE, pp 844–848
Lombrozo T, Vasilyeva N (2017) Causal explanation. In: oxford handbook of causal reasoning, Oxford University Press, Oxford, pp 415–432
Miller T (2019) Explanation in artificial intelligence: insights from the social sciences. Artif Intell 267:1–38
Mitchell T (1978) Version spaces: an approach to concept learning. Technical report, Stanford University, Department of Computer Science, Stanford
Možina M, Žabkar J, Bratko I (2007) Argument based machine learning. Artif Intell 171(10):922–937
Muggleton S, De Raedt L (1994) Inductive logic programming: theory and methods. J Logic Program 19:629–679
Muggleton S, Schmid U, Zeller C, Tamaddoni-Nezhad A, Besold T (2018) Ultra-strong machine learning: comprehensibility of programs learned with ilp. Mach Learn 107(7):1119–1140
Najarian K, Splinter R (2005) Biomedical signal and image processing. CRC Press, Boca Raton
Pesapane F, Volonté C, Codari M, Sardanelli F (2018) Artificial intelligence as a medical device in radiology: ethical and regulatory issues in Europe and the United States. Insights Imaging 9(5):745–753
Rabold J, Deininger H, Siebers M, Schmid U (2019) Enriching visual with verbal explanations for relational concepts—combining LIME with Aleph. In: Advances in interpretable machine learning and artificial intelligence workshop (AIMLAI) at ECML 2019
Samek W, Binder A, Montavon G, Lapuschkin S, Müller K-R (2016) Evaluating the visualization of what a deep neural network has learned. IEEE Trans Neural Netw Learn Syst 28(11):2660–2673
Schmid U (2018) Inductive programming as approach to comprehensible machine learning. In: Proceedings of the 6th workshop KI & Kognition (KIK-2018), co-located with KI 2018, volume http://ceur-ws.org/Vol-2194/schmid.pdf
Siebers M, Schmid U (2019) Please delete that! Why should I? Explaining learned irrelevance classifications of digital objects. KI-Künstliche Intelligenz 33(1):35–44
Siebers M, Schmid U, Seuß D, Kunz M, Lautenbacher S (2016) Characterizing facial expressions by grammars of action unit sequences-a first investigation using abl. Inf Sci 329:866–875
Srinivasan A (2001) The Aleph manual
Srinivasan A, Muggleton S, King RD, Sternberg MJ (1994) Mutagenesis: Ilp experiments in a non-determinate biological domain. In: Proceedings of the 4th international workshop on inductive logic programming, Citeseer, vol 237, pp 217–232
Ware M, Frank E, Holmes G, Hall M, Witten IH (2001) Interactive machine learning: letting users build classifiers. Int J Hum Comput Stud 55(3):281–292
Weitz K, Hassan T, Schmid U, Garbas J-U (2019) Deep-learned faces of pain and emotions: elucidating the differences of facial expressions with the help of explainable AI methods. tm-Technisches Messen 86(7–8):404–412
Winston PH (1975) Learning structural descriptions from examples. In: Winston P (ed) The psychology of computer vision. McGraw-Hil, New York, pp 157–210
Wittekind C (2016) TNM: Klassifikation maligner Tumoren. Wiley, Amsterdam
Funding
Open Access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
The work presented in this paper is part of the BMBF ML-3 project Transparent Medical Expert Companion (TraMeExCo), FKZ 01IS18056 B, 2018–2021.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schmid, U., Finzel, B. Mutual Explanations for Cooperative Decision Making in Medicine. Künstl Intell 34, 227–233 (2020). https://doi.org/10.1007/s13218-020-00633-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13218-020-00633-2