Abstract
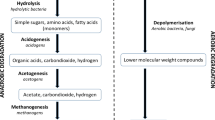
Microbial carbohydrate-active enzymes (CAZyme) can be harnessed for valorization of Lignocellulosic biomass (LCB) to value-added chemicals/products. The two Indian Rivers Ganges and the Yamuna having different origins and flow, face accumulation of carbon-rich substrates due to the discharge of wastewater from adjoining paper and pulp industries, which could potentially contribute to the natural enrichment of LCB utilizing genes, especially at their confluence. We analyzed CAZyme diversity in metagenomic datasets across the sacred confluence of the Rivers Ganges and Yamuna. Functional annotation using CAZyme database identified a total of 77,815 putative genes with functional domains involved in the catalysis of carbohydrate degradation or synthesis of glycosidic bonds. The metagenomic analysis detected ~ 41% CAZymes catalyzing the hydrolysis of lignocellulosic biomass polymers- cellulose, hemicellulose, lignin, and pectin. The Beta diversity analysis suggested higher CAZyme diversity at downstream region of the river confluence, which could be useful niche for culture-based studies. Taxonomic origin for CAZymes revealed the predominance of bacteria (97%), followed by archaea (1.67%), Eukaryota (0.63%), and viruses (0.7%). Metagenome guided CAZyme diversity of the microflora spanning across the confluence of Ganges-Yamuna River, could be harnessed for biomass and bioenergy applications.




Similar content being viewed by others
Availability of data and materials
The raw sequencing data have been deposited at NCBI under the project accession number PRJNA716890. The samples raw data were deposited under NCBI SRA accession number SRX10447184, SRX10447183, SRX10447182, SRX10447181, SRX10447180, SRX10447179, SRX10447178, respectively.
References
Al-Masaudi S, El Kaoutari A, Drula E et al (2017) A metagenomics investigation of carbohydrate-active enzymes along the gastrointestinal tract of Saudi sheep. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00666
Andrade AC, Fróes A, Lopes FÁC et al (2017a) Diversity of microbial carbohydrate-active enZYmes (CAZYmes) associated with freshwater and soil samples from Caatinga Biome. Microb Ecol 74:89–105. https://doi.org/10.1007/s00248-016-0911-9
Ausland C, Zheng J, Yi H et al (2021) dbCAN-PUL: a database of experimentally characterized CAZyme gene clusters and their substrates. Nucleic Acids Res 49:D523–D528. https://doi.org/10.1093/nar/gkaa742
Barrett K, Jensen K, Meyer AS et al (2020) Fungal secretome profile categorization of CAZymes by function and family corresponds to fungal phylogeny and taxonomy: example Aspergillus and Penicillium. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-61907-1
Bertucci M, Calusinska M, Goux X et al (2019) Carbohydrate hydrolytic potential and redundancy of an anaerobic digestion microbiome exposed to acidosis, as uncovered by metagenomics. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00895-19
Bhatia SK, Jagtap SS, Bedekar AA et al (2020) Recent developments in pretreatment technologies on lignocellulosic biomass: effect of key parameters, technological improvements, and challenges. Bioresour Technol 300:122724
Buchfink B, Xie C, Huson DH (2014) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60
Cantarel BI, Coutinho PM, Rancurel C et al (2009) The Carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233. https://doi.org/10.1093/nar/gkn663
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Carroll A, Somerville C (2009) Cellulosic biofuels. Annu Rev Plant Biol 60:165–182. https://doi.org/10.1146/annurev.arplant.043008.092125
Chettri D, Verma AK, Verma AK (2020) Innovations in CAZyme gene diversity and its modification for biorefinery applications. Biotechnol Rep 28:e00525
Choudhury KSM, Sagar A, Satish Kumar S (2018) Central pollution control board ministry of environment, forest & climate change. www.cpcb.nic.in
Chuzel L, Ganatra MB, Rapp E et al (2018) Functional metagenomics identifies an exosialidase with an inverting catalytic mechanism that defines a new glycoside hydrolase family (GH156). J Biol Chem 293:18138–18150. https://doi.org/10.1074/jbc.RA118.003302
Comtet-Marre S, Parisot N, Lepercq P et al (2017) Metatranscriptomics reveals the active bacterial and eukaryotic fibrolytic communities in the rumen of dairy cow fed a mixed diet. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00067
CPCB (Central Pollution Control Board) (2013) Pollution Assessment : River Ganga. Cent Pollut Control Board, Minist Environ For Govt India 1–206
De Coster W, D’Hert S, Schultz DT et al (2018) NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. https://doi.org/10.1093/bioinformatics/bty149
Doi RH, Kosugi A (2004) Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol 2:541–551
Dwivedi S, Chauhan PS, Mishra S et al (2020) Self-cleansing properties of Ganga during mass ritualistic bathing on Maha-Kumbh. Environ Monit Assess 192:1–15. https://doi.org/10.1007/s10661-020-8152-2
Escuder-Rodríguez J-J, DeCastro M-E, Cerdán M-E et al (2018) Cellulases from Thermophiles Found by Metagenomics. Microorganisms 6:66. https://doi.org/10.3390/microorganisms6030066
Garber AI, Zehnpfennig JR, Sheik CS et al (2021) Metagenomics of Antarctic marine sediment reveals potential for diverse chemolithoautotrophy. mSphere. https://doi.org/10.1128/MSPHERE.00770-21
Garron ML, Henrissat B (2019) The continuing expansion of CAZymes and their families. Curr Opin Chem Biol 53:82–87
Gong G, Zhou S, Luo R et al (2020) Metagenomic insights into the diversity of carbohydrate-degrading enzymes in the yak fecal microbial community. BMC Microbiol 20:302. https://doi.org/10.1186/s12866-020-01993-3
Helbert W, Poulet L, Drouillard S et al (2019) Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc Natl Acad Sci U S A 116:6063–6068. https://doi.org/10.1073/pnas.1815791116
Jin M, Guo X, Zhang R et al (2019) Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome. https://doi.org/10.1186/s40168-019-0675-9
Kameshwar AKS, Ramos LP, Qin W (2019) CAZymes-based ranking of fungi (CBRF): an interactive web database for identifying fungi with extrinsic plant biomass degrading abilities. Bioresour Bioprocess 6:51. https://doi.org/10.1186/s40643-019-0286-0
Lahti L, Salojärvi J, Salonen A et al (2014) Tipping elements in the human intestinal ecosystem. Nat Commun 5:1–10. https://doi.org/10.1038/ncomms5344
Lairson LL, Henrissat B, Davies GJ, Withers SG (2008) Glycosyl transferases: structures, functions, and mechanisms. Annu Rev Biochem 77:521–555
Ma C, Lo PK, Xu J et al (2020) Molecular mechanisms underlying lignocellulose degradation and antibiotic resistance genes removal revealed via metagenomics analysis during different agricultural wastes composting. Bioresour Technol 314:123731. https://doi.org/10.1016/j.biortech.2020.123731
Menzel P, Ng KL, Krogh A (2016) Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. https://doi.org/10.1038/ncomms11257
Montella S, Ventorino V, Lombard V et al (2017) Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci Rep 7:1–15. https://doi.org/10.1038/srep42623
Ndeh D, Rogowski A, Cartmell A et al (2017) Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544:65–70. https://doi.org/10.1038/nature21725
Pinard D, Mizrachi E, Hefer CA et al (2015) Comparative analysis of plant carbohydrate active enZymes and their role in xylogenesis. BMC Genomics. https://doi.org/10.1186/s12864-015-1571-8
Puentes-Téllez PE, Salles JF (2020) Dynamics of abundant and rare bacteria during degradation of lignocellulose from sugarcane biomass. Microb Ecol 79:312–325. https://doi.org/10.1007/s00248-019-01403-w
Reddy B, Pandey J, Dubey SK (2019) Assessment of environmental gene tags linked with carbohydrate metabolism and chemolithotrophy associated microbial community in River Ganga. Gene 704:31–41. https://doi.org/10.1016/j.gene.2019.04.004
Rittmann BE (2008) Opportunities for renewable bioenergy using microorganisms. Biotechnol Bioeng 100:203–212
Sadaiappan B, Prasannakumar C, Subramanian K, Subramanian M (2019) Metagenomic data of vertical distribution and abundance of bacterial diversity in the hypersaline sediments of Mad Boon-mangrove ecosystem, Bay of Bengal. Data Br 22:716–721. https://doi.org/10.1016/j.dib.2018.12.028
Samson R, Shah M, Yadav R et al (2019) Metagenomic insights to understand transient influence of Yamuna River on taxonomic and functional aspects of bacterial and archaeal communities of River Ganges. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2019.04.166
Samson R, Rajput V, Shah M et al (2020) Deciphering taxonomic and functional diversity of fungi as potential bioindicators within confluence stretch of Ganges and Yamuna Rivers, impacted by anthropogenic activities. Chemosphere 252:126507. https://doi.org/10.1016/j.chemosphere.2020.126507
Serrano-Ruiz JC, Luque R, Sepúlveda-Escribano A (2011) Transformations of biomass-derived platform molecules: from high added-value chemicals to fuels via aqueous-phase processing. Chem Soc Rev 40:5266–5281. https://doi.org/10.1039/c1cs15131b
Sidar A, Albuquerque ED, Voshol GP et al (2020) Carbohydrate binding modules: diversity of domain architecture in amylases and cellulases from filamentous microorganisms. Front Bioeng Biotechnol 8:871
Singhvi M, Gokhale D (2013) Biomass to biodegradable polymer (PLA). RSC Adv 3:13558–13568
Singhvi MS, Gokhale DV (2019) Lignocellulosic biomass: hurdles and challenges in its valorization. Appl Microbiol Biotechnol 103:9305–9320
Singhvi M, Kim BS (2020) Current developments in lignocellulosic biomass conversion into biofuels using nanobiotechology approach. Energies 13:5300
Stewart RD, Auffret MD, Warr A et al (2018) Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun. https://doi.org/10.1038/s41467-018-03317-6
Strazzulli A, Cobucci-Ponzano B, Iacono R et al (2020) Discovery of hyperstable carbohydrate-active enzymes through metagenomics of extreme environments. FEBS J 287:1116–1137. https://doi.org/10.1111/FEBS.15080
Sukumaran RK, Christopher M, Kooloth-Valappil P et al (2021) Addressing challenges in production of cellulases for biomass hydrolysis: targeted interventions into the genetics of cellulase producing fungi. Bioresour Technol 329:124746. https://doi.org/10.1016/j.biortech.2021.124746
Tran PQ, Bachand SC, McIntyre PB et al (2021) Depth-discrete metagenomics reveals the roles of microbes in biogeochemical cycling in the tropical freshwater Lake Tanganyika. ISME J 157(15):1971–1986. https://doi.org/10.1038/s41396-021-00898-x
Van Meerbeek K, Muys B, Hermy M (2019) Lignocellulosic biomass for bioenergy beyond intensive cropland and forests. Renew Sustain Energy Rev 102:139–149
Wang L, Zhang G, Xu H et al (2019) Metagenomic analyses of microbial and carbohydrate-active enzymes in the rumen of holstein cows fed different forage-to-concentrate ratios. Front Microbiol. https://doi.org/10.3389/fmicb.2019.00649
Wang H, Bier R, Zgleszewski L et al (2020) Distinct distribution of archaea from soil to freshwater to estuary: implications of archaeal composition and function in different environments. Front Microbiol 11:2641. https://doi.org/10.3389/FMICB.2020.576661
Xie R, Wang Y, Chen Q et al (2020) Coupling between carbon and nitrogen metabolic processes mediated by coastal microbes in synechococcus-derived organic matter addition incubations. Front Microbiol 11:1041. https://doi.org/10.3389/fmicb.2020.01041
Zhang H, Yohe T, Huang L et al (2018) DbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. https://doi.org/10.1093/nar/gky418
Zhao R, Summers ZM, Christman GD et al (2020) (2020) Metagenomic views of microbial dynamics influenced by hydrocarbon seepage in sediments of the Gulf of Mexico. Sci Reports 101(10):1–13. https://doi.org/10.1038/s41598-020-62840-z
Acknowledgements
The authors are thankful to the Directors of CSIR-NCL, CSIR-NEERI for infrastructure and support. VR acknowledges CSIR-NCL for the fellowship support through NCIM technical services project. RS, RKY acknowledge CSIR and UGC respectively for the fellowship and AcSIR for academic support.
Funding
This work is the outcome of the National Mission for Clean Ganga (NMCG) program (GKC-01/2016-17, 212, NMCG- Research) under the Ministry of Water Resources, River Development and Ganga Rejuvenation, New Delhi, India.
Author information
Authors and Affiliations
Contributions
All the authors have read and review the manuscript for submission. VR: analysis and writing original draft; RS: writing, editing, sample collection and sequencing RKY: editing and sequencing; MD: supervision, designing study, and reviewing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rajput, V., Samson, R., Yadav, R. et al. Metagenomic mining of Indian river confluence reveal functional microbial community with lignocelluloytic potential. 3 Biotech 12, 132 (2022). https://doi.org/10.1007/s13205-022-03190-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03190-7