Abstract
Symptoms typical of phytoplasma infection such as phyllody, virescence, witches’ broom and yellowing were observed in 12 varieties of Chrysanthemum morifolium in floral nurseries and experimental fields at New Delhi, Karnataka, Maharashtra and Andhra Pradesh, India, during surveys made from 2015 to 2017. Disease incidence ranged from 15 to 30%. Phytoplasma presence was confirmed in all symptomatic chrysanthemum varieties by molecular identification assays. Sequence comparison, phylogenetic and in silico RFLP analyses of 16S rDNA sequences allowed the identification of the chrysanthemum infecting phytoplasma strains into different ribosomal groups and subgroups, namely 16SrI, 16SrII-D, 16SrVI-D and 16SrXIV. Detection of phytoplasma strains of 16SrII-D subgroup were also confirmed in symptomatic Chenopodium album and Parthenium hysterophorus plants grown in and around the surveyed chrysanthemum fields at New Delhi, whereas 16SrVI-D phytoplasma strains were detected in symptomatic Cannabis sativa weed and leafhopper Hishimonus phycitis individuals collected from the symptomatic chrysanthemum fields at New Delhi. This is the first report on the presence of 16SrVI and 16SrXIV groups of phytoplasmas in chrysanthemum plants. Studies on genetic diversity of phytoplasmas infecting the major chrysanthemum varieties in India and their epidemiological aspects had previously not been reported. The detection and identification of phytoplasmas in different chrysanthemum varieties could contribute to increase the awareness among farmers in the management of these diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chrysanthemum (Chrysanthemum morifolium Ramat) occupies 3rd and 5th positions in the cut flower and pot plant trades; respectively (Anonymous 2015). It is used as cut flower and loose flower due to its attractive form and colour of flower. In India, it is grown commercially on a large area of 16.63 thousand ha with a production of 186.06 thousand MT (Anonymous 2014). Flower crops are affected worldwide by several biotic and abiotic stresses and phytoplasma-associated diseases are the major threat to commercial cultivations and are responsible of severe economic losses (Chaturvedi et al. 2010; Bellardi et al. 2018).
The occurrence of phytoplasmas in chrysanthemum has been reported from Japan, Italy, China, Iran and India (Shiomi and Sugiura 1983; Conti et al. 1988; Raj et al. 2007; Chung 2008; Min et al. 2009; Rani et al. 2014; Yadav et al. 2015). Although chrysanthemum has been grown at a commercial scale in different states of India, very little is known about the phytoplasma strains infecting this crop (Rao et al. 2017). Therefore, the genetic diversity of phytoplasmas infecting the major chrysanthemum varieties in India was studied. In addition, molecular identification of these phytoplasmas presence in weeds that may act as alternative hosts and potential insect vectors responsible of their spread were also made.
Materials and methods
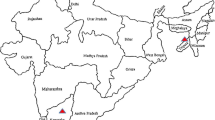
Surveys of gardens, nurseries and experimental fields at ICAR-Indian Agricultural Research Institute (ICAR-IARI), New Delhi, ICAR-Indian Institute of Horticultural Research (IIHR), Bangalore, Karnataka, Maharashtra (Bandra and Pune) and Andhra Pradesh (Rajahmundry) were made from October 2015 to February 2017 (Fig. 1). Symptoms of phytoplasma infections were recorded on 12 chrysanthemum varieties from 5 surveyed locations. The disease incidence on each variety was evaluated on the basis of visual inspection by counting the number of symptomatic plants in relation to the total number of plants inspected using formula:
Three leaf samples each of symptomatic chrysanthemum varieties from all the four locations and three weed species (Chenopodium album, Parthenium hysterophorus and Cannabis sativa) from IARI, New Delhi were collected (Table 1). Toria phyllody phytoplasma (Azadvar et al. 2009) maintained in periwinkle (Catharanthus roseus) in the greenhouse was used as a positive control.
Asymptomatic chrysanthemum plant samples were collected from the fields of the same area but at distance from the symptomatic fields with no symptoms and used them as negative controls. Leafhopper species from the surveyed chrysanthemum fields at New Delhi were also collected at 15 days intervals from October 2016 to February 2017 by the sweeping net method. The captured leafhoppers were identified by morphology as per taxonomy manual on identification of leafhoppers (Viraktamath and Meshram 2019) at Division of Entomology, ICAR- Indian Agricultural Research Institute, New Delhi.
DNA from plants and insects was extracted employing CTAB-based extraction procedures as described (Ahrens and Seemüller 1992; Maixner et al. 1995). PCR amplification was performed with the universal phytoplasma primer pair P1/P7 (Deng and Hiruki 1991; Schneider et al. 1995) followed by primer pair 3Far/3Rev (Manimekalai et al. 2010). PCR reactions were carried out following the protocol described by Rao et al. (2014). Five microlitres of PCR product was subjected to electrophoresis in a 1.0% (w/v) agarose gel, stained with ethidium bromide and observed under a UV transilluminator. All PCR products (~ 1.3 kb) were purified using the Wizard® SV Gel and PCR Clean- up System (Promega, Madison, USA).
PCR products were ligated in pGEM®T vector (Promega) and cloned in Escherichia coli (DH5-α) following the manufacturer’s instructions, the cloned products were outsourced for sequencing using M13Fwd/M13Rev universal primer pair in both directions at Agrigenome, Kerala, India. The sequences were assembled using DNA baser V.4 software online tool and submitted to GenBank. A database search was performed by BLASTn analysis at NCBI (www.ncbi.org). The 16S rRNA gene sequences were aligned with phytoplasma group/subgroup representatives available in GenBank using ClustalW (Thompson et al. 1994) and used to construct a phylogenetic tree by the Neighbor-joining method with 1000 replications for each bootstrap value using MEGA 7.0 software (Kumar et al. 2016). The 16Sr DNA sequence (Acc. No. AB680603) of Acholeplasma laidlawii was used as out group to root the phylogenetic tree.
Results
During survey of gardens, nurseries, experimental fields in four states of India, symptoms of leaf yellowing, phyllody, virescence, witches’ broom and stunting were recorded on chrysanthemum varieties. The phyllody and witches’ broom symptoms were recorded as most common symptoms (Figs. 2, 3). The symptoms recorded on chrysanthemum varieties in four states of India are listed in Table 1. The disease incidence on different chrysanthemum varieties were recorded from 15% in Indira variety at Kadiam, Rajahmundry, Andhra Pradesh to 30% in Pusa Anmol variety at IARI, New Delhi (Table 1). Symptoms were most pronounced at the flowering stage at all the surveyed locations.
Phytoplasma disease symptoms in different chrysanthemum varieties at IARI, New Delhi: a PusaKesari: phyllody and virescence symptoms; b Pusa centenary: witches’ broom and flower virescence symptoms; c Ajay Orange: severe phyllody and witches’ broom; d Pusa Anmol: severe virescence and phyllody; e Red Spoon: witches’ broom and phyllody; f Jaya: phyllody
Phytoplasma disease symptoms in different chrysanthemum varieties a Johan Webber: severe phyllody at IARI New Delhi; b Indira: leaf yellowing and severe stunting at Kadiam, Rajahmundry; c White Nursery: yellowing and stunting at DFR, Pune; d Nayak Yellow: leaf curling and yellowing at DFR, Pune; e Meera: greening of flowers at Bandra; f Yellow Gold: necrosis and witches’ broom at IIHR, Bangalore
Symptoms including leaf chlorosis, phyllody and witches’ broom were also observed in Chenopodium album, Parthenium hysterophorus and Cannabis sativa grown in and around the chrysanthemum fields at ICAR-IARI, New Delhi (Table 1, Fig. 4). Among the weed species, P. hysterophorus showed the highest disease incidence (14.5%) followed by C. album (12.60%) and C. sativa (11.20%) (Table 1).
All the symptomatic chrysanthemum and weed plants (Table 1) along with toria phyllody phytoplasma strain yielded amplicons of ~ 1.3 kb in nested PCR assays. No amplification products were obtained from the template DNA from any asymptomatic plants with each of primer pair employed. Leafhoppers collected from the surveyed chrysanthemum experimental fields at IARI, New Delhi were identified as Hishimonus phycitis, Empoasca motti, Hecalus porrectus, Amrasca biguttula and Psammotettix spp. Among these species, H. phycitis was recorded as the most abundant species.
Amplification products were obtained from individuals of H. phycitis but not from those of the other four leafhopper species (Empoasca motti, Hecalus porrectus, Amrasca biguttula, Psammotettix spp) collected from the field at IARI, New Delhi (data not shown).
A BLASTn identity search of GenBank database revealed that phytoplasma strains infecting chrysanthemum plants of varieties Pusa Centenary (GenBank Acc. No.KY612250), Ajay Orange (GenBank Acc. Nos. KX641012, KY693690), Pusa Anmol (GenBank Acc. Nos. KY693687, KY693688), Red Spoon (GenBank Acc. No. MF040140) and Indira from Rajahmundry, Andhra Pradesh (GenBank Acc. No.KY693693), all from IARI, New Delhi showed 16S rDNA sequence similarity 98.5 to 100% with 16SrII phytoplasma group (Table 1). Phytoplasma strains detected in chrysanthemum plants of varieties Jaya (GenBank Acc. No. KY693689) and Johan Webber (GenBank Acc. No. KX641014), both from IARI, New Delhi, and White Nursery (GenBank Acc. No. KY693691) from DFR, Pune showed 16S rDNA sequence similarity 99.4–99.8% with ‘Candidatus Phytoplasma trifolii’ 16SrVI group. However, the phytoplasma strains associated with Pusa Kesari from IARI, New Delhi (GenBank Acc. No. KX641015), Yellow Gold from IIHR, Bangalore (GenBank Acc. No.KY693692), and Meera from Bandra, Mumbai (GenBank Acc. No. KY472314) showed sequence similarity ranging from 99.7 to 100% with 16SrI group (Table 1), whereas phytoplasma strains from chrysanthemum Nayak Yellow (GenBank Acc. No. KX641016) from IARI, New Delhi, hosted a phytoplasma strain which showed 16S rDNA sequence similarity 99.6% with 16SrXIV group (Table 1).
Phytoplasma strains detected in C. album (GenBank Acc. No. MF040139) and P. hysterophorus (GenBank Acc. No.MF040141) plants showed a 99.9–100% 16S rDNA sequence identity with phytoplasma described in peanut witches’ broom group whereas those present in C. sativa plants and H. phycitis leafhopper showed a 99.9–100% 16S rDNA sequence identity with 16SrVI group.
Phylogenetic analysis of 16S rDNA sequences showed that phytoplasma strains detected in different varieties of chrysanthemum, weed plants and H. phycitis leafhoppers were clustered with phytoplasmas enclosed in 16SrI, 16SrII, 16SrVI and 16SrXIV groups (Fig. 5). Subgroup assignment by comparison of virtual RFLP patterns derived from in silico digestions 17 restriction endonucleases indicated Pusa Centenary (GenBank Acc. No. KY612250), Ajay orange strain 1 (GenBank Acc. No. KX641012) Ajay orange strain 2 (GenBank Acc. No. KY693690), Pusa anmol strain1 (GenBank Acc. No KY693687) and Pusa anmol strain 2 (GenBank Acc. No KY693688) Red spoon (GenBank Acc. No. MF040140), Chenopodium album (GenBank Acc. No. MF040139) and Parthenium hysterophorus (GenBank Acc. No. MF040141) belonged to 16SrII subgroup-D, However the RFLP pattern of phytoplasma strain Indira (GenBank Acc. No. KY693693) with the reference strain papaya mosaic disease phytoplasma (GenBank Acc. No. Y10096) showed variation for enzymes AluI, BfaI, HaeIII and HpaI could not be assigned to subgroups because of variation in the RFLP patterns with reference strain (Fig. 6a, Table 1). Jaya (GenBank Acc. No. KY693689), White Nursery (GenBank Acc. No. KY693691), Cannabis sativa (GenBank Acc. No. MF509775) and H. phycitis phytoplasma strain (GenBank Acc. No KY856746) belonged to 16SrVI subgroup-D. However, the RFLP pattern of phytoplasma strain Johan webber (GenBank Acc. No. KX641014) with the reference strain periwinkle little leaf phytoplasma (GenBank Acc. No. AF228053) showed variation for enzymes EcoRI, MboI, RsaI and TaqI and could not be assigned to subgroups because of variation in the RFLP patterns with reference strain of 16SrVI-D subgroup (Fig. 7, Table 1). The RFLP pattern of phytoplasma strain Pusa Kesari (GenBank Acc. No. KX641015) showed variation for enzymes AluI, BstUI, HaeIII and MseI; Yellow gold (GenBank Acc. No. KY693692) showed variation for enzymes AluI, BfaI, HaeIII, HlnfI, HpaII and RsaI and Meera (GenBank Acc. No KY472314) showed variation for enzymes, BstUI and HpaII with reference strain of 16SrI-B subgroup (GenBank Acc. No. M30790) could not be assigned to subgroups because of variation in the RFLP patterns with reference strain (Fig. 8, Table 1). The Nayaka yellow (GenBank Acc. No. KX641016) belonged to 16SrXIV group but could not assigned to subgroup because of variation in profilings of Mse1 and Taq1 (Fig. 9, Table 1).
Phylogenetic tree based on 16Sr DNA constructed by neighbor-joining method showing the relationship among chrysanthemum varieties, weed species and insect phytoplasma strains, and reference phytoplasma strains. Accession numbers are specified in the tree. ‘Acholeplasma laidlawaii’ was used as outgroup. Mega 7.0 software was used to construct the tree. Numbers on branches are bootstrap values obtained for 1000 replicates. The bar represents a phylogenetic distance of 0.01
Comparison of virtual RFLP patterns derived from in silico digestions of ~ 1.3 kb 16S rDNA sequences of a Reference strain 16SrII-D Mollicutes sp. (associated with papaya mosaic disease) 16S rRNA gene (GenBank Acc. No. Y10096); b Pusa Centenary (GenBank Acc. No. KY612250); c Ajay orange strain 1 (GenBank Acc. No. KY641012); d Ajay orange strain 2 (GenBank Acc. No. KY693690; e Pusa anmol strain1(GenBank Acc. No KY693687; f Pusa anmol strain 1(GenBank Acc. No KY693688; g Red Spoon (GenBank Acc. No. MF040140); h Indira (GenBank Acc. KY693693); i Chenopodium album (GenBank Acc No. MF040139); j Parthenium hysterophorus (GenBank Acc. No. MF040141) digested using 17 different restriction endonucleases indicating that chrysanthemum varieties phytoplasma belonged to 16SrII-D phytoplasma sub groups, but the Indira strain showed variation for the RFLP pattern
Comparison of virtual RFLP patterns derived from in silico digestions of ~ 1.3 kb 16S rDNA sequences of a Reference strain 16SrVI-D Periwinkle little leaf phytoplasma 16S rRNA gene. (GenBank Acc. No. AF228053), b Jaya (GenBank Acc. No. KY693689) c White Nursery (GenBank Acc. No. KY693691, d Cannabis sativa (GenBank Acc. No. MF509775) e Hishimonas phycitis phytoplasma (GenBank Acc. No KY856746) and f Johan Webber (GenBank Acc. No. KX641014) digested using 17 different restriction endonucleases indicating that Jaya, White Nursery, Cannabis sativa and Hishimonas phycitis phytoplasma belonged to 16SrVI-D sub groups, but Johan Webber 'Chrysanthemum showed variation for RFLP pattern and could not be assigned subgroup
Comparison of virtual RFLP patterns derived from in silico digestions of ~ 1.3 kb 16S rDNA sequences of a Reference strain Oenothera phytoplasma (GenBank Acc. No. M30790) and b Pusa Kesari phytoplasma (GenBank Acc. No. KX641015) c Yellow gold phytoplasma (GenBank Acc. No. KY693692) and d Meera phytoplasma (GenBank Acc. No. KY472314) digested using 17 different restriction endonucleases indicating variations for RFLP pattern. Hence subgroup could not be assigned
Comparison of virtual RFLP patterns derived from in silico digestions of ~ 1.3 kb 16S rDNA sequences of a Reference strain 16SrXIV-A, Bermuda grass white leaf phytoplasma 16S rRNA gene (GenBank Acc. No. AJ550984), b Nayaka yellow (GenBank Acc. No. KX641016) digested using 17 different restriction endonucleases indicating that this strain belonged to 16SrXIV group but could not be assigned subgroup 16SrXIV-A subgroup because of difference in MseI and Taq1 restriction profiles
Discussion
Different groups of phytoplasmas are reported earlier with different chrysanthemum species. Symptoms induced by aster yellows phytoplasma infection included stunt, yellowing, virescence and phyllody in C. frutescence in Italy (Bertaccini et al. 1992; Conti et al. 1988). The presence of phytoplasmas affiliated to aster yellows group in chrysanthemum was reported earlier from Italy, Korea and India (Marzachi and Bosco 2005; Chung 2008; Raj et al. 2007). Chrysanthemum yellows phytoplasma (16SrI-B subgroup) associated with witches’ broom, vein clearing, dwarf and abnormal production of secondary shoots was reported in C. carinatum and C. morifolium from European and Mediterranean areas (Bertaccini et al. 1990; Conti et al. 1988; Saracco et al. 2005). The identification of 16SrII group phytoplasmas in C. grandiflorum was reported in Okinawa Prefecture, Japan (Naito et al. 2007) and India (Yadav et al. 2015). Min et al. (2009) reported a new outbreak of phytoplasma disease of chrysanthemum with symptoms of yellowing of leaf margins, flattened stem and shortening of internodes in China associated with 16SrI group, which was responsible for serious losses in flower quality.
Limited reports are available on identification of phytoplasma strains association with chrysanthemum varieties in India (Raj et al. 2007; Yadav et al. 2015). However, in the present study, two subgroups belonging to four groups of phytoplasmas (16SrI, 16SrII-D, 16SrVI-D and 16SrXIV) infecting 12 varieties of chrysanthemum were identified from four major chrysanthemum growing states of India. However, the restriction profiles of phytoplasma strain associated with chrysanthemum variety Johan weber from Delhi (Acc. No. KX641014), Nayak yellow from Pune (Acc. No. KX641016), Meera from Mumbai (Acc. No. KY472314) and Indira from Andhra Pradesh did not match completely with any of the earlier classified subgroups, hence they may be new strains and multilocus genes specific detection and real RFLP analysis are necessary to confirm the reported unusual variability.
Several weeds are reported as reservoirs of important phytoplasma strains which are suggested to play an important role in natural spread of phytoplasma strains and serve as natural alternative hosts, since they influence the population density of the vectors and act as source of inoculums (Pasquini et al. 2007; Duduk et al. 2018). In the present study, two phytoplasmas ribosomal groups (16SrII and 16SrVI) were identified in three weed species, that are also reported to be reservoir of different phytoplasma strains in different countries, viz., 16SrII-E subgroup in Italy (Tolu et al. 2006), 16SrIII (X-disease) group and 16SrXII group in Czech Republic (Safarova et al. 2011). In the present study, the report of Chenopodium sp. as a host of 16SrII group indicates its potentiality as natural reservoir. Earlier, P. hysterophorus weed was also reported as host of 16SrI and 16SrII group in India and China (Raj et al. 2009; Li et al. 2011; Mall et al. 2015; Yadav et al. 2016). Further, C. sativa and Achyranthes aspera weed species were also reported as host of 16SrI, 16SrII, 16SrVI and 16SrXIV groups of phytoplasmas in India (Rao et al. 2017), which suggests potentiality of these weed species as natural reservoirs of phytoplasmas. C. album, P. hysterophorus and C. sativa weed species that are prevalent in chrysanthemum fields at New Delhi and resulted positive for the 16SrII and 16SrVI groups of phytoplasmas in the present study may also play an important role for natural spread of phytoplasma strains.
Limited information is available on transmission of phytoplasma strains associated with ornamentals through insect vectors. Bosco et al. (2007) reported transmission of chrysanthemum yellows phytoplasma through three leafhopper vector species (Euscelis incisus, Euscelidius variegatus and Macrosteles quadripunctulatus) in Italy. Empoasca decipiens was proved to be an experimental vector in transmitting chrysanthemum yellows phytoplasma (16SrI) in Italy (Galetto et al. 2011a; b). However, in the present study, the leafhopper, H. phycitis was positive to16SrVI group phytoplasmas. The identification of 16SrVI group of phytoplasma strain in H. phycitis and in two chrysanthemum varieties, Jaya and Johan Webber, in same field suggest it as vector, which needs further confirmation through transmission assays. This leafhopper species has already been reported as vectors for 16SrI and 16SrII phytoplasma groups in India (Nabi et al. 2015; Gopala and Rao 2018), which indicates its potentiality to transmit different phytoplasma strains. In India, many crops are being cultivated in parallel with other agricultural crops in different seasons. The widespread occurrence of phytoplasma strains identified in chrysanthemum varieties in the study pose a serious threat of chrysanthemum cultivation in India and needs further studies on epidemiology and management.
References
Ahrens U, Seemüller E (1992) Detection of DNA of plant pathogenic mycoplasma like organisms by a polymerase chain reaction that amplifies a sequence of the 16S rRNA gene. Phytopathology 82:828–832
Anonymous (2014) NHB database. Indian Horticulture Database, Gurgaon, Haryana. Ministry of Agriculture, Government of India. https://nhb.gov.in
Anonymous (2015) Flora Holland.Legmeerdijk 313, Aalsmeer, Netherlands. https://www.floraholland.com
Azadvar M, Baranwal VK, Yadava DK (2009) First report of a 16SrIX (Pigeon pea witches’ broom) phytoplasma associated with toria (Brassica rapa cv. toria) phyllody disease in India. New Dis Rep 20:27
Bellardi MG, Betaccini A, Madhupriya RGP (2018) Phytoplasma disease in ornamental crops. In: Rao GP, Bertaccini A, Fiore N, Liefting LW (eds) Phytoplasmas: plant pathogenic bacteria-I, characterization and epidemiology of phytoplasma-associated diseases. Springer, Singapore, pp 191–234
Bertaccini A, Davis RE, Lee I-M, Conti M, Dally EL, Douglas SM (1990) Detection of chrysanthemum yellows mycoplasma like organism by dot hybridization and Southern blot analysis. Plant Dis 74:40–43
Bertaccini A, Davis RE, Hammond RW, Bellardi MG, Vibio M, Lee IM (1992) Sensitive detection of mycoplasma like organisms in field collected and in vitro propagated plants of Brassica, Hydrangea and Chrysanthemum by polymerase chain reaction. Ann Appl Biol 121(3):593–599
Bosco D, Galetto L, Leoncini P, Saracco P, Raccah B, Marzachì C (2007) Interrelationships between ‘CandidatusPhytoplasma asteris’ and its leafhopper vectors (Homoptera: Cicadellidae). Econ Entomol 100:1504–1511
Chaturvedi Y, Singh M, Snehi SK, Raj SK, Rao GP (2010) First report of ‘Candidatus Phytoplasma asteris’ (16Sr I group) associated with yellows and little leaf diseases of Hibiscus rosa-sinensis in India. Plant Pathol 59(4):796–796
Chung BN (2008) Phytoplasma detection in Chrysanthemum and lily. In: Harrison NA, Rao GP, Marcone C (eds) Characterization, diagnosis and management of phytoplasma. Studium Press LLC, Texas, pp 175–194
Conti M, Agostino GD, Casetta A, Mela L (1988) Some characteristics of Chrysanthemum yellows disease. Acta Hortic 234:129–136
Deng S, Hiruki C (1991) Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. J Microbiol Methods 14(1):53–61
Duduk B, Stepanovi`c J, Yadav A, Rao GP (2018) Phytoplasma in weeds and wild plants. In: Rao GP, Bertaccini A, Fiore N, Liefting LW (eds) Phytoplasmas: plant pathogenic bacteria-I, characterisation and epidemiology of phytoplasma-associated diseases. Springer, Singapore, pp 288–313
Galetto L, Bosco D, Balestrini R, Genre A, Fletcher J, Marzachì C (2011a) The major antigenic membrane protein of “Candidatus Phytoplasma asteris” selectively interacts with ATP synthase and actin of leafhopper vectors. PLoS ONE 6:7
Galetto L, Marzachì C, Demichelis S, Bosco D (2011b) Host plant determines the phytoplasma transmission competence of Empoasca decipiens (Hemiptera: Cicadellidae). J Econ Entomol 104(2):360–366
Gopala, Rao GP (2018) Molecular characterization of phytoplasma associated with four important ornamental plant species in India and identification of natural potential spread sources. 3 Biotech 8(2):116
Kumar S, Stecher G, Tamura K (2016) MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Li Z, Zhang L, Che H, Liu H, Chi M, Luo D, Li Y, Chen W, Wu Y (2011) A disease associated with phytoplasma in Parthenium hysterophorus. Phytoparasitica 39:407–410
Maixner M, Ahrens U, Seemüller E (1995) Detection of the German grapevine yellows (Vergilbungskrankheit) MLO in grapevine, alternative hosts and a vector by a specific PCR procedure. Eur J Plant Pathol 101:241–250
Mall S, Kumar S, Jadon VJ, Rao GP (2015) Identification of phytoplasmas associated with weed species in India. Indian Phytopathol 68(4):449–453
Manimekalai R, Soumya VP, Sathish KR, Selvarajan R, Reddy K, Thomas GV, Sasikala M, Rajeev G, Baranwal VK (2010) Molecular detection of 16SrXI group phytoplasma associated with root (wilt) disease of coconut (Cocos nucifera) in India. Plant Dis 94:636
Marzachí C, Bosco D (2005) Relative quantification of chrysanthemum yellows (16SrI) phytoplasma in its plant and insect host using real-time polymerase chain reaction. Mol Biotech 30(2):117–127
Min H, Hu SB, Li ZN, Wu YF, Zhang CP, Wei T (2009) A phytoplasma associated with an outbreak of an unusual disease of Chrysanthemum in China in 2008. Plant Dis 93(8):840–840
Naito T, Tanaka M, Taba S, Toyosato T, Oshiro A, Takaesu K, Hokama K, Usugi T, Kawano S (2007) Occurrence of chrysanthemum virescence caused by “Candidatus Phytoplasma aurantifolia” in Okinawa. J Gen Plant Pathol 73(2):139–141
Pasquini G, Ferretti L, Barba M (2007) Diffusione del legnonerodellavitenel Lazio e caratterizzazione molecolare dell’ agenteeziologico. Inf Fitopatol 4:42–47
Raj SK, Khan MS, Kumar S (2007) Molecular identification of Candidatus Phytoplasma asteris associated with little leaf disease of Chrysanthemum morifolium. Australas Plant Dis Notes 2:21–22
Raj SK, Snehi SK, Kumar S, Banerji BK, Dwivedi AK, Roy RK, Goel AK (2009) First report of ‘Candidatus phytoplasma asteris’ (16SrI group) associated with colour-breaking and malformation of floral spikes of gladiolus in India. Plant Pathol 58:1170
Rani A, Misra P, Singh J, Kumar P, Rani R, Shukla (2014) PCR based detection of phytoplasma association in pot marigold (Calendula officinalis L.) and guldawari (Dendranthema grandiflora L.). Asian J Biol Sci 9(2):238–241
Rao GP, Tiwari AK, Kumar S, Baranwal VK (2014) Identification of sugarcane grassy shoot-associated phytoplasma and one of its putative vectors in India. Phytoparasitica 42:349–354
Rao GP, Madhupriya TV, Manimekalai R, Tiwari AK, Yadav A (2017) A century progress of research on phytoplasma diseases in India. Phytopathogenic Mollicutes 7(1):1–38
Safarova D, Valova P, Flidr P, Navratil M (2011) Molecular identification of 16SrIII and 16SrXII phytoplasma groups in Chenopodium album in Czech Republic. Bull Insectol 64:85–86
Saracco P, Bosco D, Veratti F, Marzachì C (2005) Quantification over time of chrysanthemum yellows phytoplasma (16Sr-I) in leaves and roots of the host plant Chrysanthemum carinatum (Schousboe) following inoculation with its insect vector. Physiol Mol Plant Pathol 67(3–5):212–219
Schneider B, Seemüller E, Smart CD, Kirkpatrick BC (1995) Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasmas. In: Razin S, Tully JG (eds) Molecular and diagnostic procedures in mycoplasmology, vol 1. Academic Press, San Diego, pp 369–380
Shiomi T, Sugiura M (1983) Water dropwort yellows and Chrysanthemum witches’ broom occurred in Ishikawa Prefecture. Jpn J Phytopathol 49(3):367–370
Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW- improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tolu G, Botti S, Garau R, Prota VA, Sechi A, Prota U, Bertaccini A (2006) Identification of a 16SrII-E Phytoplasma in Calendula arvensis, Solanumnigrum, and Chenopodium spp. Plant Dis 90(3):325–330
un Nabi S, Dubey DK, Rao GP, Baranwal VK, Sharma P (2015) Molecular characterization of ‘Candidatus Phytoplasma asteris’ subgroup IB associated with sesame phyllody disease and identification of its natural vector and weed reservoir in India. Australas Plant Pathol 44(3):289–297
Viraktamath CA, Meshram NM (2019) Leafhopper tribe Coelidiini (Hemiptera: Cicadellidae: Coelidiinae) of the Indian subcontinent. Zootaxa. https://doi.org/10.11646/zootaxa.4653.1.1
Yadav V, Mahadevakumar S, Sreenivasa MY, Janardhana GR (2015) First report on the occurrence of virescence of chrysanthemum associated with 16SrII-A group phytoplasma in India. Plant Dis 99(11):1641
Yadav A, Thorat V, Shouche Y (2016) Candidatus Phytoplasma aurantifolia (16SrII Group) Associated with Witches’ Broom Disease of Bamboo (Dendrocalamus strictus) in India. Plant Dis 100(1):209
Acknowledgements
Authors wish to thank Director, IARI, New Delhi for providing lab facilities and Head, Division of Floriculture and Landscaping, IARI for help in field survey work and recording disease incidence in ornamentals fields at IARI, New Delhi.
Author information
Authors and Affiliations
Contributions
AT carried out survey, collected the samples, did the PCR assays for plants and insect samples and also edited and submitted the sequence in GenBank. DVSR, NB and GK helped in survey of floral nurseries for the incidence of phytoplasma disease and collected of weeds and insects in chrysanthemum fields at IARI, New Delhi. PP helped in analysing the sequences for the identification of phytoplasma strains. RM helped in pDraw analyses of sequence data. CM and GPR helped in writing and editing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
GenBank submission
All the 16Sr RNA gene sequences has been submitted in GenBank and appeared in public database.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Taloh, A., Raju, D.V.S., Banyal, N. et al. Genetic diversity of phytoplasma strains infecting chrysanthemum varieties in India and their possible natural reservoirs. 3 Biotech 10, 411 (2020). https://doi.org/10.1007/s13205-020-02407-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02407-x