Abstract
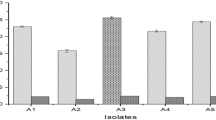
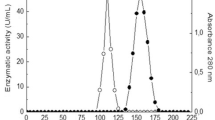
The tannase production ability by endophytic actinobacteria and the genetic identity of responsible tannase gene were determined. The studied strains were isolated from surface-sterilized leaf discs of Ailanthus excelsa Roxb. Four strains were found to hydrolyze tannic acid on solid media containing 0.4% tannic acid. The strain AL1L was found as tan B LP indicating production of tannase with diverse of substrate affinity. The tannase production from the potential strain AL1L was performed in liquid tannic acid broth (0.4%, w/v). The strain was later identified as Streptomyces sp. AL1L on the basis of 16S rDNA homology. Highest enzyme activity was observed at 48 h of incubation at the exponential growth phase. The enzyme was purified by ammonium sulfate precipitation followed by dialysis (15 kD cut off). This enzyme, with molecular weight 180 kD shows highest catalytic activity at 35 °C, pH 6 with substrate concentration 0.1 g%. The purified enzyme possesses 1.4 × 10−3 K m and 11.15 U/ml as V max. The above study indicates high industrial prospective of endophytic actinobacteria as source of tannase of potential biotechnological applications.




Similar content being viewed by others
References
Aguilar CN, Rodríguez R, Gutiérrez-Sánchez G, Augur C, Favela-Torres E, Prado-Barragan LA, Ramírez-Coronel A, Contreras-Esquivel JC (2007) Microbial tannases: advances and perspectives. Appl Microbiol Biotechnol 76:47–59
Banerjee D, Mahapatra S (2012) Fungal tannase: a journey from strain isolation to enzyme application. Dyn Biochem Process Biochem Mol Biol 6:49–60
Banerjee D, Pati BR (2007) Optimization of tannase production by Aureobasidium pullulans DBS66. J Microbiol Biotechnol 17:1049–1053
Chae S, Yu T (1983) Experimental manufacture of a com wine by fungal tannase. Hanguk Sipkum Kwahakoechi 15:326–332
Chaitanyakumar A, Anbalagan M (2016) Expression, purification and immobilization of tannase from Staphylococcus lugdunensis MTCC 3614. AMB Express 6:89. https://doi.org/10.1186/s13568-016-0261-5
Curiel JA, Rodríguez H, Acebrón I, Mancheño JM, de las Rivas B, Muñoz R (2009) Production and physicochemical properties of recombinant Lactobacillus plantarum tannase. J Agric Food Chem 57:6224–6230. https://doi.org/10.1021/jf901045s
Farias GM, Gorbea C, Elkins JR, Griffin GJ (1994) Purification characterization and substrate relationships of the tannase from Cryphonectria parasitica. Physiol Mol Plant Pathol 44:51–63
Gordon RE, Barnett DA, Handerhan JE, Pang CHN (1974) Nocardia coeliaca, Nocardia autotrophica and the Nocardin strain. Int J Syst Evol Microbiol 24:54–63. https://doi.org/10.1099/00207713-24-1-54
Hatamoto O, Watarai T, Kikuchi M, Mizusawa K, Sekine H (1996) Cloning and sequencing of the gene encoding tannase and a structural study of the tannase subunit from Aspergillus oryzae. Gene 175:215–221
Iwamoto K, Tsuruta H, Nishitaini Y, Osawa R (2008) Identification and cloning of a gene encoding tannase (tannin acylhydrolase) from Lactobacillus plantarum. ATCC 14917T. Syst Appl Microbiol 31:269–277. https://doi.org/10.1016/j.syapm.2008.05.004
Jiménez N, Curiel JA, Reverón I, de las Rivas B, Muñoz R (2013) Uncovering the Lactobacillus plantarum WCFS1 gallate decarboxylase involved in tannin degradation. Appl Environ Microbiol 79:4253–4263. https://doi.org/10.1128/AEM.00840-13
Jiménez N, Esteban-Torres M, Mancheño JM, de las Rivas B, Muñoz R (2014) Tannin degradation by a novel tannase enzyme present in some Lactobacillus plantarum strains. Appl Environ Microbiol 80:2991–2997
Khanbabaee K, Ree TV (2001) Tannins: classification and definition. Nat Prod Rep 18:641–649
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lekha PK, Lonsane BK (1997) Production and application of tannin acyl hydrolase: state of the art. In: Neidleman S, Laskin A (eds) Advances in Applied microbiology, vol 44. Academic, San Diego, pp 215–260
Li J, Qiong X, Yufeng H, Hui N, Changzheng W, Anfeng X (2017) Tannase application in secondary enzymatic processing of inferior Tieguanyin oolong tea. Electron J Biotechnol 28:87–94
Liu TPSL, Costa RMPB, Freitas DJV, Nacimento CO, Motta CMS, Bezerra RP, Herculano PN, Porto ALF (2017) Tannase from Aspergillus melleus improves the antioxidant activity of green tea: purification and biochemical characterization. Int J Food Sci Technol 52(3):652–661
Lu MJ, Chu SC, Yan L, Chen C (2009) Effect of tannase treatment on protein-tannin aggregation and sensory attributes of green tea infusion. LWT Food Sci Technol 42:338–342
Noguchi N, Ohashi T, Shiratori T, Narui K, Hagiwara T, Ko M, Watanabe K, Miyahara T, Taira S, Moriyasu F, Sasatsu M (2007) Association of tannase-producing Staphylococcus lugdunensis with colon cancer and characterization of a novel tannase gene. J Gastroenterol 42:346–351. https://doi.org/10.1007/s00535-007-2012-5
Ow YY, Stupans I (2003) Gallic acid and gallic acid derivatives: effects on drug metabolizing enzymes. Curr Drug Metab 4:241–248
Ramirez-Coronel MA, Viniegra-Gonzalez G, Darvill A, Augur C (2003) A novel tannase from Aspergillus niger with β-glucosidase activity. Microbiology 149:2941–2946
Ren B, Wu M, Wang Q, Peng X, Wen H, McKinstry WJ, Chen Q (2013) Crystal structure of tannase from Lactobacillus plantarum. J Mol Biol 425:2737–2751. https://doi.org/10.1016/j.jmb.2013.04.032
Rodríguez-Durán LV, Valdivia-Urdiales B, Contreras-Esquivel JC, Rodríguez-Herrera RPK, Aguilar CN (2011) Novel strategies for upstream and downstream processing of tannin acyl hydrolase. Enzyme Res 2011:823619. https://doi.org/10.4061/2011/823619
Roy S, Banerjee D (2015a) Bioactive endophytic actinobacteria of Cinnamomum sp.; isolation, identification, activity guided purification and process optimization of active metabolite. Am J Microbiol 6:4–13. https://doi.org/10.3844/ajmsp.2015
Roy S, Banerjee D (2015b) Broad spectrum antibacterial activity of granaticinic acid, isolated from Streptomyces thermoviolaceus NT1; an endophyte in Catharanthus roseus (L.) G. Don. J Appl Pharm Sci 5:6–11. https://doi.org/10.7324/JAPS.2015.50702
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sharma KP, John PJ (2011) Purification and characterization of tannase and tannase gene from Enterobacter sp. Process Biochem 46:240–244. https://doi.org/10.1016/j.procbio.2010.08.016
Sivashanmugam K, Jayaraman G (2011) Production and partial purification of extracellular tannase by Klebsiella pneumoniae MTCC 7162 isolated from tannery effluent. Afr J Biotechnol 10:1364–1374. https://doi.org/10.5897/AJB10.1209
Thorington RW Jr, Ferrell KE (2006) Squirrels: the animal answer guide. Johns Hopkins University Press, Baltimore, p 91 (ISBN 0-8018-8402-0)
Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA (1983) Numerical classification of Streptomyces and related genera. Microbiology 129:1743–1813. https://doi.org/10.1099/00221287-129-6-1743
Wu M, Wang Q, McKinstry WJ, Ren B (2015) Characterization of a tannin acyl hydrolase from Streptomyces sviceus with substrate preference for digalloyl ester bonds. Appl Microbiol Biotechnol 99:2663–2672. https://doi.org/10.1007/s00253-014-6085-9
Zhong X, Peng L, Zheng S, Sun Z, Ren Y, Dong M, Xu A (2004) Secretion purification and characterization of a recombinant Aspergillus oryzae tannase in Pichia pastoris. Protein Expr Purif 36:165–169
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roy, S., Parvin, R., Ghosh, S. et al. Occurrence of a novel tannase (tan B LP ) in endophytic Streptomyces sp. AL1L from the leaf of Ailanthus excelsa Roxb. 3 Biotech 8, 33 (2018). https://doi.org/10.1007/s13205-017-1055-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-1055-4