Abstract
Micronutrient deficiencies are of major concern in human health and plant metabolism. Iron (Fe), zinc (Zn), iodine (I), selenium (Se) are regarded as micronutrients having major impact on human health. More than 50% of populations mainly from developing countries are suffering from one or the other micronutrient malnutrition. Ensuring adequate supply of these micronutrients through diet consisting of staple foods, such as common bean (Phaseolus vulgaris L.) is must. Here, we evaluated common bean genotypes that were collected from various regions of Jammu and Kashmir, India for Fe, Zn and protein contents and used SSRs to identify the markers associated with these traits. We found significant variation among genotypes for Fe, Zn and protein contents. Genotype R2 was having 7.22 mg 100 g−1 of Fe content, genotype K15 with 1.93 mg 100 g−1 of Zn content and genotype KS6 with 31.6% of protein content. Diversity study was done using both cluster and structure based approach. Further, association mapping analysis using General Linear Method (GLM) approach was done to identify SSRs associated with accumulation of Fe, Zn and protein. 13 SSRs were identified that significantly (p < 0.05) showed association with Fe, Zn and protein contents in common bean. The markers associated with Fe were located on chromosome no. 2, 5, 6, 7, 9 and 10, markers associated with Zn were located on chromosome no. 1, 3, 5, 7 and 10 whereas only one marker located on chromosome no. 4 was found associated with protein content. These findings will provide potential opportunity to improve Fe and Zn concentrations in common bean, through molecular breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common bean (Phaseolus vulgaris L.) is one of the most important legume crop widely used as staple food throughout the world (Broughton et al. 2003). It is cultivated in many parts of world as dry beans and green beans (Ibarra-Perez et al. 1997; Burle et al. 2010). The sufficient amount of proteins (20–28%), carbohydrates, vitamins, dietary fiber (56%), micronutrients e.g., iron (Fe) (70 mg/kg) and zinc (Zn) (33 mg/kg), vitamin B complex (niacin, riboflavin, folic acid and thiamine) and polyunsaturated fatty acids present in common bean have made it a potential staple food to overcome the problem of hunger (Pennington and Young 1990; Welch et al. 2000; Broughton et al. 2003; Guzman and Infante 2007). The deficiency of Fe, Zn, I, Se and Vitamin A are responsible for 65% of childhood deaths. About 80% of the global population is affected by Fe deficiency and 30% of the individuals mainly from developing countries are suffering from anemia (Welch and Graham 2004). It is also reported that approximately 50% of the world’s population is affected by Zn deficiency. As such both Fe and Zn deficiencies are the major problems faced by public health sector in the world (Gebremedhin et al. 2011). To address the micronutrient deficiencies, it is important to enhance these micronutrients in various staple food items (Zhang et al. 2012). Enhancement of micronutrients in staple food crops through breeding is one of the most promising approaches. So far various studies have been conducted to analyze variation in mineral concentrations in common bean which is a prerequisite for improvement through breeding (Welch and Graham 2004; Talukder et al. 2010; Tryphone and Nchimbi-Msolla 2010; Mahajan et al. 2015). Till date, information on the genetic bases for various important traits (seed quality, disease resistance, mineral concentration and other morpho-phenological characters) in common bean were derived from linkage and quantitative trait locus (QTL) analysis using segregating populations. Various QTLs linked to different traits in common bean includes resistance to disease (Frei et al. 2005; Blair et al. 2006a), yield related traits (Blair et al. 2006b), drought tolerance (Blair et al. 2012a, b), mineral concentration (Guzma´n-Maldonado et al. 2003; Cichy et al. 2005; Gelin et al. 2006; Cichy et al. 2008; Blair et al. 2010a, b) are known. However, it is worth mentioning here that QTL analysis using biparental population requires two or more generations and very large segregating populations for a fine resolution map. Moreover, the markers identified are often specific to cross or pedigree and only two alleles at any particular locus can be studied simultaneously (Flint-Garcia et al. 2003). Association mapping study emerges as a powerful approach for mapping economically and biologically valuable traits using unstructured diverse germplasm. This approach is based on linkage disequilibrium (LD), which refers to the non random association of alleles at different loci (Lewontin and Kojima 1960). It is a most commonly used alternative approach to bi-parental mapping for the identification of genes responsible for natural variation in phenotypic/biochemical or other traits of interest. Through association mapping, it is now possible to map the chromosomal location of genes/QTLs controlling traits of interest at a much greater resolution. Association mapping has been used in many major crops to identify markers associated with a particular trait, such as wheat (Breseghello and Sorrells 2006; Jaiswal et al. 2012), maize (Thornsberry et al. 2001; Wilson et al. 2004; Camus-Kulandaivelu et al. 2006; Salvi 2007), rice (Bao et al. 2006; Agrama et al. 2007; Zhao et al. 2011), barley (Kraakman et al. 2006), sorghum (Casa et al. 2008; Shehzad et al. 2009), potato (Malosetti et al. 2007), sugarcane (Wei et al. 2006) and soybean (Jun et al. 2007). Some association mapping studies in common bean for various traits such as common bacterial blight (CBB) (Shi et al. 2011), yield components (Galeano et al. 2012), agronomical traits (Nemli et al. 2014), nutrient elements (Nemli et al. 2016) have also been reported. In the present investigation, we used a diverse core set of 96 common bean genotypes representing a collection from Jammu & Kashmir India for mapping of QTLs contributing for the accumulation of seed Fe, Zn and protein. A set of 55 polymorphic SSRs covering all the chromosomes were used for genotyping and association analysis. 13 SSRs were identified which showed association with Fe, Zn and protein content in common bean.
Materials and methods
Genotypes
A diverse core set of 96 genotypes out of a collection of 138 common bean genotypes consisting of mainly landraces collected from various unexploited regions of Jammu and Kashmir and one check line VLR-125 (Almorha, VPKAS, India) (Mahajan et al. 2017) were used as an experimental material for association mapping.
Chemical analysis
Estimation of micronutrient (Fe and Zn) and protein contents
Fe and Zn concentration of each genotype was estimated by diacid method by Jackson (1973). For the detailed procedure refer Mahajan et al. (2015). The concentration values were further converted and expressed in mg 100 g−1. Protein content was determined using Lowry’s method (Lowry et al. 1951). For the detailed procedure refer Mahajan et al. (2015). Protein content was calculated and expressed in percentage.
Marker resources
74 SSRs (Genomic and Geneic) with known positions on 11 chromosomes were used (Yu et al. 2000; Gaitan-Solis et al. 2002; Grisi et al. 2007; Hanai et al. 2010; Co´ rdoba et al. 2010). Out of 74 SSRs only 55 SSRs were polymorphic and these 55 SSRs were considered for further analysis (Supplementary Table 1).
DNA isolation, PCR, and SSR analysis
The genomic DNA was extracted from the leaf tissues using CTAB method by Doyle and Doyle (1987). The DNA quantity as well as quality was checked by Nanodrop (mySPEC, Wilmington, USA). It was diluted to a final concentration of 25 ng/μL for enabling polymerase chain reactions. PCR amplification was carried out in 96 well Universal Gradient Thermal Cycler (PEQLAB, Deutschland and Osterrtich, United Kingdom) in a 10 µL reaction mixture. The reaction mixture contained 5 µM of each forward and reverse primers, 1 U of Taq polymerase (D1806- Sigma Aldrich, USA), 5 µL of 10X PCR buffer with MgCl2, 2.5 mM of each dNTP (dTTPs, dGTPs, dCTPs, dATPs). Amplifications were performed using following program (94 °C for 4 min, followed by 30 cycles of 94 °C for 30 s, 47–56 °C for 30 s, 72 °C for 30 s, with a final extension of 72 °C for 8 min). PCR products were mixed with 6X loading dye (Thermo scientific # R0611) (2–3 µL) and resolved on 3% metaphor Agarose gel. The primers that did not give clear polymorphic pattern on metaphor Agarose were further tested on 8% denaturing Poly Acrylamide Gel Electrophesis (PAGE). PCR products were visualized either using UV light for metaphor Agarose gels (MiniLumi, Sigma-Svi Bio Solutions Pvt. Ltd. New- Delhi, India) or by silver-staining of PAGE gels (Bassam et al. 1991). The clear and reproducible alleles amplified by each SSR were scored according to their fragment size (bp) corresponding to the 50 bp molecular weight marker (10488-043-Invitrogen).
Data analysis
Biochemical data
All observations were taken in three replicates and values were then averaged. One-way ANOVA was applied to evaluate the variance of mineral contents (Fe and Zn) and protein among the genotype. The Pearson’s correlation coefficient between Fe, Zn and protein was determined by Pearson’s correlation analysis using SPSS (version 20.0).
Genotypic data
The amplified DNA fragments identified as a band in the gel were considered as a distinct phenotype and independent of the others, determining the alleles of each SSR locus. Allele differences were determined by their relative mobility in gel and the size of each allele was estimated using 50 bp DNA Ladder. When a PCR product was not amplified, data for the relevant genotype was treated as null allele. Genetic variation was estimated by calculating various parameters such as Gene diversity, Polymorphism Information Content (PIC), Heterozygosity, Major allele frequency and Number of allele score using Power Marker software version 3.25 (Liu and Muse 2005).
Diversity analysis
Phylogenetic tree was constructed using pair wise distance matrix computed by calculating a dissimilarity matrix using a shared allele index with DARwin software (Perrier and Jacquemoud-Collet 2006). An unweighted neighbor joining tree was constructed using the calculated dissimilarity index. The genetic distance between accessions was estimated using NEI coefficient with bootstrap procedure of resampling (1000) across markers and individuals from allele frequencies (Nei 1972). In addition, Principal Coordinate Analysis (PCoA) was done using DARwin software (Perrier and Jacquemoud-Collet 2006).
Population structure analysis
To assess population structure, Model based approach was followed using STRUCTURE ver 2.3.4 software (Pritchard et al. 2000). The actual number of subpopulation which is represented by K was identified by this method. STRUCTURE analysis was carried out with a length of burn-in of 150,000 and MCMC (Markov Chain Monte Carlo) of 150,000 with the possibility of admixture and allele frequency correlated. Ten independent simulations were conducted allowing K (number of subpopulations) to vary from 2 to 10. The optimum K value was determined by plotting the mean estimate of the log posterior probability of the data (L (K) against the given K value. True number of subpopulation was identified using the maximal value of L (K). An adhoc quantity ΔK proposed by Evano et al. (2005) based on second order rate of change of the likelihood function with respect to K estimated using Structure Harvester (Earl and vonHoldt 2012) has also shown a clear peak at the optimal K value. A line was assigned to a given cluster when the proportion of its genome in the cluster (qK) was higher than a standard threshold value of 50%. For the chosen optima value of K, membership coefficient matrices of replicates from STRUCTURE were integrated to generate a Q matrix using the software CLUMPP (Jakobsson and Rosenberg 2007) and the STRUCTURE bar plot was drawn using the STRUCTURE PLOT software (Ramasamy et al. 2014).
Marker trait association
The concentration of Fe, Zn and protein contents in common bean seeds were subjected to association analysis with SSR loci of 96 diverse genotypes in common bean seeds. The association analysis between the marker and traits of interest were performed based on the General Linear Model (GLM) using the software TASSEL 3.0 (Bradbury et al. 2007). Genotypic microsatellite data with Fe, Zn and protein content were combined for identifying markers associated with these traits. Markers were considered to be associated with the traits if the markers show significant (p < 0.05) association.
Results and discussion
Fe, Zn and protein contents
The seed material of all the common bean genotypes collected from various regions of Jammu & Kashmir, India were analysed for Fe, Zn and protein contents. The results obtained were further statistically analysed. The mean ± SE for each observation is presented in supplementary Table 2.
Wide variation was observed in seed Fe concentration ranging from 0.59 to 7.22 mg 100 g−1 with an average of 1.42 mg 100 g−1. Genotype R2 possesses highest seed Fe content (7.22 mg 100 g−1) whereas genotype UG7 had lowest Fe content (0.59 mg 100 g−1). The Zn concentration in seed varied from 0.15 to 1.93 mg 100 g−1 with an average of 0.70 mg 100 g−1. Genotype K15 possesses highest Zn content in seed (1.93 mg 100 g−1) whereas genotype R1 had lowest Zn content (0.15 mg 100 g−1). Supplementary Table 2 and Supplementary Figure 1 represent the mean values of Fe and Zn content observed in seeds of 138 common bean genotypes. In earlier reports similar pattern of variation in Fe and Zn content in common bean seeds has been reported. A wide variation in Fe and Zn contents among 100 diverse common bean lines ranging from 54.20 to 161.50 and 29.33 to 65.50 mg kg−1, respectively, has been observed (Silva et al. 2010). Similarly, variation in Fe and Zn content from 34 to >100 and 21 to 54 mg kg−1, respectively, among 2000 common bean accession of CIAT has been observed (Beebe et al. 2000). In a recent report, 117 genotypes of common bean collected from Uganda showed variation in Fe and Zn contents ranging from 45 to 87 and 22 to 40 mg kg−1, respectively (Mukamuhirwa et al. 2012). Since, there is a huge variation in Fe and Zn contents in our material; it is worth to mention here that the germplasm can act as a potential genetic resource for improving nutritional quality of adopted common bean cultivars, through breeding.
Further, the seeds were analysed for their protein content and a wide variation was observed ranging from 7.2 to 31.6% with an average of 18.83%. Highest protein content was observed in KS6 (31.6%) whereas genotype K12 had the least protein content of 7.2%. Supplementary Table 2 and Supplementary Figure 1 represent the mean values of protein content observed in the seeds of 138 diverse common bean genotypes. Similar results have been reported in previous studies, although variation may be influenced by environmental factors, geographical location and growing season. In earlier reports, protein content in the range of 17.4–29% (Sood et al. 2003; Silva and Iachan 1975; Sgarbieri et al. 1979; Marquez and Lajolo 1981; Durigan and Sgarbieri 1985; Durigan et al. 1987; Tezoto and Sgarbieri 1990) has been recorded. Protein content of 36 North American bean cultivars was evaluated and content ranged from 19.6 to 32.2% (Koehler et al. 1987). Wide range in micronutrients (Fe and Zn) and protein contents among 138 genotypes in this study, indicates the existence of extensive genetic variation which can be explored for improving the nutritional quality of this crop. Further, these 138 genotypes were grouped on the basis of their mean values (Supplementary Table 3) to cluster them based on particular range of micronutrient (Fe and Zn) and protein contents.
Correlation among mineral (Fe and Zn) and protein contents
The Pearson correlation analysis of mineral and protein contents of 138 diverse genotypes showed that there exists a non-significant weak positive correlation among Fe, Zn and Protein contents. Fe and Zn (r = 0.101; p > 0.05), Fe and Protein (r = 0.040; p > 0.05) and Zn and Protein (r = 0.034; p > 0.05) (Table 1; Supplementary Figure 1). Our results confirmed earlier reports, where Fe and Zn, Fe and Protein, Zn and Protein content in common bean seeds were found non-significantly and positively correlated, r = 0.14; r = 0.25; r = 0.40; p > 0.05, respectively (Mario et al. 2009). So, it can be interpreted that accumulation of one mineral has little impact on the concentration of the other, as such there is a genetic regulation governing the transport and accumulation of these micronutrients.
Allele diversity
The results obtained from 55 polymorphic SSRs are summarized in Table 2. These 55 SSRs were found to be functional as they amplified PCR product of expected size range. A total of 396 alleles ranging from 2 (PVBR 5 and X79722) to 20 (X74919) were amplified on 96 genotypes with an average of 7.2 alleles per loci (Table 2). Earlier studies revealed varying number of amplified alleles per loci in common bean. 7.14 alleles per loci were observed in set of 29 common bean accessions (Maras et al. 2008). 7 alleles per loci in common bean landraces of Brazil (Burle et al. 2010). An average of 5.7 alleles per loci was reported in Nicaragua genotypes (Gome´z et al. 2004). However, an average of 8.9 alleles per loci were reported in Nicaragua genotypes (Jime´nez and Korpelainen 2012), an average of 10 alleles per loci were reported in two different common bean genotype collections from East (Asfaw et al. 2009) and Central Africa (Blair et al. 2010a, b) and 19 alleles per loci in Uganda genotypes (Okii et al. 2014). Depending on number of genotypes and SSRs used in genotyping, the number of alleles varies accordingly.
The utility of a marker is a balance between the level of polymorphism it can detect, and its capacity to identify multiple alleles (Powell et al. 1996). We observed, PIC value ranging from 0.214 (BMb1008) to 0.832 (BM 150) with an average of 0.6128 (Table 2). This indicates the informativeness of markers used in the present study. Further, gene diversity was calculated. It is the probability that two randomly chosen alleles from the population are different. For the material used in present study, the gene diversity value varied from 0.2278 (BMb1008) to 0.8508 (BM 150) with an average value of 0.6611 (Table 2). Gene diversity study conducted earlier in other common bean population ranged from 0.07 to 0.83 with the mean value of 0.50 (McClean et al. 2012). The major allele frequency in the present study ranged from 0.1957 (BM150) to 0.8737 (Bmb1008) with an average of 0.4531 for all SSR markers (Table 2). Only 29% of SSR markers were found to have value of more than 0.50 which means that the major allele in most of the markers is not shared by most of the common bean germplasm used in the present study. Earlier study conducted also showed similar mean value of major allele frequency in different common bean genotypes. An average of 0.47 major allele frequency was reported in 349 wild, landraces and commercial varieties collected from Latin American, Europe, USA, Africa, and Asia (Kwak and Gepts 2009). An average of 0.31 major allele frequency among the common bean genotypes was reported (Okii et al. 2014). However, we also calculated heterozygosity, to have information about the proportion of heterozygous individuals in the population. In this study the value of heterozygosity ranged from 0.000 (PVBR5, BMb1008, PVBR182, X79722, PVBR14, BMd10) to 0.873 (BM155) with an average of 0.22. In earlier studies, higher mean heterozygosity was observed. Mean heterozygosity of 0.45 with highest heterozygosity value of 0.96 was reported (Okii et al. 2014). Thus, the differences in the major allele frequency, PIC and heterozygosity suggest that these discriminatory parameters are strongly influenced by the number and diversity of the genotypes under evaluation and may be due to the difference in number of polymorphic markers used.
Genetic diversity
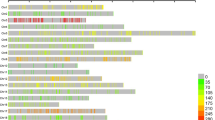
Further cluster analysis of 96 diverse genotypes was done. The pair-wise distance between all possible pairs of 96 diverse genotypes was calculated to identify genetic relationship among them. The dendrogram obtained divided common bean genotypes into three main clusters namely I, II and III (Fig. 1). Most of the genotypes grouped in individual clusters belonged to a particular region. Cluster I is further divided into a major and minor cluster. Minor cluster includes only four genotypes two from Baramulla, one from Bandipora and Rajouri whereas the major cluster which is further divided into sub sub cluster includes all of the genotypes collected from Ugada, Kashmir, Nishant, Marmat, Udhampur, Desa, Marwa, most of the genotypes from Bhaderwah and few from Poonch. Cluster II is also divided into two sub cluster which includes most of the genotypes from Rajouri, Bandipora and Shopian and few from Poonch. However, cluster III includes most of the genotypes collected from Poonch, two from Bhaderwah and one from each Baramulla and Qazigund. Dissimilarity coefficient values were used to identify most similar and dissimilar group of genotypes. The genotypes B1 and B2 from Bhaderwah were most similar with a dissimilarity coefficient value of 0.209 and the genotypes K14 and UD3 from Bandipora and Udhampur, respectively, formed the most diverse pair of genotypes with a value of 0.809. Principal Component Analysis revealed significant diversity in this germplasm collected. The germplasm exhibiting the maximum number of population (k = 1) showed very distinct PCoA plot, population 1 concentrated only in quadrant 3 and 4, population 2 mainly concentrated in quadrant 3, population 3 concentrated in 1 and 4 quadrant, population 4 is mainly concentrated in quadrant 2, whereas population 5 is in quadrant 1 (Supplementary Figure 2).
Population structure analysis
STRUCTURE analysis was carried out to observe the number of populations from 96 genotypes using 55 SSR markers. A total of 5 sub-populations were observed from 96 diverse common bean genotypes (Fig. 2). Each individual was assigned to one of the five sub-population based on the membership probability >0.5 and the individual with probability lower than 0.50 was represented as admixture (Supplementary Table 4). Maximum number of individuals (23) were observed in sub-population 1 (Table 3). The Evanno test found a clear maximum for Delta K at K = 5 in the plots of L (K) versus Delta (Fig. 3) confirming a likely assignment of the bean germplasm to five sub-groups. The individual membership coefficient at K = 5 from the STRUCTURE run had maximum mean probability of likelihood value of L (K) = −11,248.93 which led to assignment of common bean germplasm to five (K = 5) sub populations (Fig. 4). The five sub-populations had moderate differentiation, with F ST value ranging from 0.1745 to 0.2766, mean value of 0.2246 and the expected heterozygosity which measures the probability that two randomly chosen individual will be different (heterozygous) at given locus ranged from 0.5232 to 0.5960 with a mean value of 0.5647 (Table 4). The genetic differentiation based on F ST values between five common bean sub-populations identified by population structure analysis is given in Table 5. In some of the earlier studies F ST values 0.27 (Asfaw et al. 2009) and 0.203 (Blair et al. 2012a, b) in sub-population of common bean germplasm have been observed. The difference in the F ST values may be due to the use of different types of markers in our study. Diversity in common bean germplasm was analysis for ecological distribution using cluster and structure analysis. However, tight association could not be established between structure, traits and ecological groups. On the whole, two separate analyses done in the present study basically agreed with each other having few deviations. The minor differences between groupings was due to two different methods, the cluster analysis assigned a fixed branch position to each accession and structure analysis resulted in a sub-population membership percentage, and the highest percentage was used to assign individuals to groups for easy interpretation.
Graphical representation of the optimal number of groups in the program STRUCTURE inferred using the criterion of Evano et al. (2005). The analysis was based on data obtained from 55 microsatellite loci in common bean collection evaluated for genetic diversity
Plots of L (K) against the likely sub populations (K) generated according to Evano et al. (2005) with sub-populations in Jammu and Kashmir common bean with five sub populations (K = 5)
Marker-trait association
Association mapping was carried out to estimate the degree to which gene/marker and trait or phenotype are associated/linked together in a particular population on the basis of linkage disequilibrium (Zondervan and Cardon 2004). In the present study, association mapping was done to identify markers associated with mineral and protein contents. Using General Linear Model (GLM), thirteen SSR markers were identified associated with Fe, Zn and protein content at the p < 0.05 probability level, and contributing 12–33% of the phenotypic variation under consideration (Table 6). However, further confirmation may be required for better understanding of correlation and its utilization. It was observed that, out of thirteen markers, seven markers (BM154, BM157, BM156, PVBR93, BM185, PVBR113, and Bmb742) were associated with Fe accumulation whereas 5 markers (BM157, PVBR69, M99479, Bmd803 and BM200) were found associated with Zn accumulation and only one marker (J04555) was associated with protein content (Table 6). Of thirteen markers, only one marker (BM157) was associated with more than one trait which showed significant R 2 value for multiple traits such as Fe (0.34) and Zn (0.33) suggesting significant association for Fe and Zn accumulation. Thus, this marker could be considered as multi-trait MTAs markers (Table 6). Earlier in a bi-parental population of G14519 × G4825, a set of QTLs for Fe and Zn were discovered on linkage group b06 suggesting a possibly pleiotropic locus and common physiology for mineral uptake or loading (Blair et al. 2010a, b). The trait-associated SSR markers identified in the present study, once validated can be used eventually for marker-assisted genetic improvement of common bean. The markers associated with Fe were found to be located on chromosome no. 2, 5, 6, 7, 9 and 10. The marker markers associated with Zn were found located on chromosome no. 1, 3, 5, 7 and 10, and the only marker associated with protein content was found located on chromosome no. 4. Earlier bi-parental approach also identified the QTLs for mineral concentration in common bean. One of the marker BM185 which was identified as QTL for Fe in RIL population derived by G21242 × G21078 (Blair et al. 2011) was found to be associated with Fe in our study and this validates our results. Moreover, the associated markers identified in this study are found located on chromosomes in which earlier QTLs were detected. We found markers on chromosome 2, 6, 7 and 9 associated with Fe, chromosome no. 3, 7 for Zn. In the present study, we could identify some new markers associated with Fe, Zn and protein. New markers for Fe accumulation are identified on chromosome no. 5 and 10, marker for Zn on chromosome no. 1, 5 and 10 and only one marker on chromosome no. 4 was identified for accumulation of protein. The SNP based genome-wide association mapping using 166 common bean accessions for identifying genes associated with these traits has been reported recently (Nemli et al. 2016). To our best knowledge, these findings on trait associations using SSR markers are the first report on Indian common bean germplasm. This would act as an important genetic resource for improving nutritional quality of common bean varieties through molecular breeding and provide an opportunity to the breeders in selecting markers for downstream experimentation towards crop improvement in common bean and related legumes.
Conclusion
In the present study, evaluation of common bean for Fe, Zn and protein contents was done. The results revealed wide variability in Fe, Zn and protein content in the genetic stock used in this study. Weak positive correlation among Fe, Zn and protein contents was observed. Population STRUCTURE analysis divided the common bean germplasm into 5 sub-populations. Further, for the first time, our finding showed significant association of markers for Fe, Zn and protein content in common bean core collection of Jammu & Kashmir, India using 55 polymorphic SSR markers. As most of the markers are multi-allelic in nature, these markers may be proved useful for following inheritance patterns for individual QTL incorporated in backcross breeding programmes. Thirteen markers associated with three different traits were identified with the significance level p < 0.05. Some of the SSR markers identified by association mapping analysis were previously identified through QTL analysis, as such our results are validated. Association mapping studies use relatively simple methodology and only moderately examined genotypes which overrule the limitation of QTL studies. From our study, we could observe that the Chr 5 and 7 carries multiple genes that influence Fe and Zn accumulation. The identified associated markers for Fe, Zn and Protein content may be helpful for common bean geneticists and breeders, in selection of traits and further molecular breeding for nutritional improvements of this crop.
References
Agrama HA, Eizenga GC, Yan W (2007) Association mapping of yield and its components in rice cultivars. Mol Breed 19:341–356
Asfaw A, Blair MW, Almekinders C (2009) Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African highlands. Theor Appl Genet 120:1–12
Bao JS, Corke H, Sun M (2006) Microsatellites, single nucleotide polymorphisms and a sequence tagged site in starch-synthesizing genes in relation to starch physicochemical properties in nonwaxy rice (Oryza sativa L.). Theor Appl Genet 113:1185–1196
Bassam BJ, Caetano-Anolles G, Gresshoffer PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Ann Biochem 196(1):80–83
Beebe S, Gonzalez A, Rengifo J (2000) Research on trace minerals in the common bean. Food Nutr Bull 21:387–391
Blair MW, Iriarte G, Beebe S (2006a) QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean × wild common bean (Phaseolus vulgaris L.) cross. Theor Appl Genet 112:1149–1163
Blair MW, Mun˜oz C, Garza R, Cardona C (2006b) Molecular mapping of genes for resistance to the bean pod weevil (Apion godmani Wagner) in common bean. Theor Appl Genet 112:913–923
Blair MW, Gonzales LF, Kimani PM, Butare L (2010a) Genetic diversity, inter-gene pool introgression and nutritional quality of common beans (Phaseolus vulgaris L.) from central africa. Theor Appl Genet 121:237–248
Blair MW, Medina JL, Astudillo C, Rengifo J, Beebe SE, Machado G, Graham R (2010b) QTL for seed iron and zinc concentration and content in a Mesoamerican common bean (Phaseolus vulgaris L.) population. Theor Appl Genet 121:1059–1070
Blair MW, Astudillo C, Rengifo J, Beebe SE, Graham R (2011) QTL analyses for seed iron and zinc concentrations in an intra-genepool population of Andean common beans (Phaseolus vulgaris L.). Theor Appl Genet 122:511–521
Blair MW, Galeano CH, Tovar E, Mun˜oz-Torres MC, Castrillo´n AV, Beebe SE, Rao IM (2012a) Development of a Mesoamerican intra-genepool genetic map for quantitative trait loci detection in a drought tolerant 3 susceptible common bean (Phaseolus vulgaris L.) cross. Mol Breeding 29:71–88
Blair MW, Soler A, Corte´s AJ (2012b) Diversification and population structure in common beans (Phaseolus vulgaris L.). PLoS ONE 7(11):49488
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticumaestivum L.) cultivars. Genetics 172:1165–1177
Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.) -model food legumes. Plant Soil 252:55–128
Burle ML, Fonseca JR, Kami JA, Gepts P (2010) Microsatellite diversity and genetic structure among common bean (Phaseolus vulgaris L.) landraces in Brazil, a secondary center of diversity. Theor Appl Genet 121:801–813
Camus-Kulandaivelu L, Veyrieras JB, Madur D, Combes V, FourmannM Barraud S, Dubreuil P, Gouesnard B, Manicacci D, Charcosset A (2006) Maize adaptation to temperate climate: relationship between population structure and polymorphism in the Dwarf 8 gene. Genetics 172:2449–2463
Casa AM, Pressoira G, Brown PJ, Mitchell SE, Rooney WL, Tuinstrac MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48:30–40
Cichy KA, Forster S, Grafton KF, Hosfield GL (2005) Inheritance of seed zinc accumulation in navy bean. Crop Sci 45:864–870
Cichy KA, Caldas GV, Snapp SS, Blair MW (2008) QTL analysis of seed iron, zinc, and phosphorus levels in an andean bean population. Crop Sci 49(5):1742
Co´ rdoba JM, Chavarro C, Schlueter JA, Jackson SA, Blair MW (2010) Integration of physical and genetic maps of common bean through BAC-derived microsatellite markers. BMC Genom 11:436
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure to small amounts of fresh leaf tissue. Phytochem Bull 19:11–15
Durigan JF, Sgarbieri VC (1985) Antinutritrional factors and toxicity in raw dry beans (Phaseolus vulgaris L.) of 12 Brazilian cultivars. J Food Biochem 11:185–200
Durigan JF, Sgarbieri VC, Bulisani EA (1987) Protein value of dry bean cultivars: factors interfering with biological utilization. J Agric Food Chem 35:694–698
Earl D, vonHoldt B (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Evano G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Frei A, Blair MW, Cardona C, Beebe SE, Gu H, Dorn S (2005) QTL mapping of resistance to Thrips palmi Karny in common bean (Phaseolus vulgaris L.). Crop Sci 45:379–387
Gaitan-Solis E, Duque MC, Edwards KJ, Tohme J (2002) Microsatellite repeats in common bean (Phaseolus vulgaris): isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Sci 42:2128–2136
Galeano CH, Cortés AJ, Fernández AC, Soler Á, Franco-Herrera N, Makunde G, Vanderleyden J, Blair MW (2012) Gene-based single nucleotide polymorphism markers for genetic and association mapping in common bean. BMC Genet 13:48
Gebremedhin S, Enquselassie F, Umeta M (2011) Prevalence of prenatal zinc deficiency and its association with socio-demographic, dietary and health care related factors in Rural Sidama, Southern Ethiopia: a cross-sectional study. BMC Public Health 11:898
Gelin JR, Forster S, Grafton KF, McClean PE, Rojas-Cifuentes GA (2006) Analysis of seed zinc and other minerals in a recombinant inbred population of navy bean (Phaseolus vulgaris L.). Crop Sci 47(4):1361
Gome´z O, Blair MW, Frankow-Lindberg B, Gullberg U (2004) Molecular and phenotypic diversity of common bean landraces from Nicaragua. Crop Sci 4:1412–1418
Grisi MCM, Blair MW, Gepts P, Brondani C, Pereira PAA, Brondadi RPV (2007) Genetic mapping of a new set of microsatellite markers in a reference common bean (Phaseolus vulgaris) population BAT93 × Jalo EEP558. Genet Mol Res 3:691–706
Guzma´n-Maldonado SH, Martı´nez O, Acosta-Gallegos JA, Guevara-Lara F, Paredes-Lo´pez O (2003) Putative quantitative trait loci for physical and chemical components of common bean. Crop Sci 43:1029–1035
Guzman NE, Infante JAG (2007) Antioxidant activity in cotyledon of black and yellow common beans (Phaseolus Vulgaris L.). Res J Biol Sci 2(1):112–117
Hanai LR, Santini L, Camargo LEA, Fungaro MHP, Gepts P, Tsai SM, Vieira MLC (2010) Extension of the core map of common bean with EST-SSR, RGA, AFLP and putative functional markers. Mol Breed 25(1):25–45
Ibarra-Perez F, Ehdaie B, Waines G (1997) Estimation of out crossing rate in common bean. Crop Sci 37:60–65
Jackson ML (1973) Soil Chemical Analysis. Prentice-hall of India Pvt. Ltd., New Delhi, p 498
Jaiswal V, Mir RR, Mohan A, Balyan HS, Gupta PK (2012) Association mapping for pre-harvest sprouting tolerance in common wheat (Triticumaestivum L.). Euphytica 188:89–102
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806
Jime´nez OR, Korpelainen H (2012) Microsatellite markers reveal promising genetic diversity and seed trait associations in common bean landraces (Phaseolus vulgaris L) from Nicaragua. Plant Genet Resour 10(2):108–118
Jun TH, Van K, Kim MY, Lee SH, Walker DR (2007) Association analysis using SSR markers to find QTL for seed protein content in soybean. Euphytica 162:179–191
Koehler HH, Chang CH, Scheier G, Burke DW (1987) Nutrient composition, protein quality, and sensory properties of thirty-six cultivars of dry beans (Phaseolus vulgaris L.). J Food Sci 52:1335–1340
Kraakman ATW, Martinez F, Mussiraliev B, v.Eeuwijk FA, Niks FA (2006) Linkage disequilibrium mapping of morphological, resistance, and other agronomically relevant traits in modern spring barley cultivars. Mol Breed 17:41–58
Kwak M, Gepts P (2009) Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor Appl Genet 118:979–992
Lewontin RC, Kojima K (1960) The evolutionary dynamics of complex polymorphisms. Evolution 14(4):458–472
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1):265–275
Mahajan R, Zargar SM, Aezum AM, Farhat S, Gani M, Hussain ISA, Agrawal GK, Rakwal R (2015) Evaluation of iron, zinc, and protein contents of common bean (Phaseolus vulgaris L.) genotypes: a collection from Jammu & Kashmir, India. Legume Genom Genet 6(2):1–7
Mahajan R, Zargar SM, Singh R, Salgotra RK, Farhat S, Sonah H (2017) Population structure analysis and selection of core set among common bean genotypes from Jammu and Kashmir, India. Appl Biochem Biotechnol 182(1):16–28. doi:10.1007/s12010-016-2307-1
Malosetti M, van der Linden CG, Vosman B, van Eeuwijk FA (2007) A mixed-model approach to association mapping using pedigree information with an illustration of resistance to phytophthorainfestans in potato. Genetics 175:879–889
Maras M, Susˇtar-Vozlicˇ J, Javornik B, Meglicˇ V (2008) The efficiency of AFLP and SSR markers in genetic diversity estimation and gene pool classification of common bean (Phaseolus vulgaris L.). Acta Agric Slovenica 91:87–96
Mario PC, Viviana BV, Juan TU (2009) Inorganic nutritional composition of common bean (Phaseolus vulgaris L.) genotypes race Chile. Chil J Agric Res 69(4):486–495
Marquez UML, Lajolo FM (1981) Composition and digestibility of albumin, globulins, and glutelins from Phaseolus vulgaris. J Agr Food Chem 29:1068–1074
McClean PE, Terpstra J, McConnell M, White C, Lee R, Mamidi S (2012) Population structure and genetic differentiation among the USDA common bean (Phaseolus vulgaris L.) core collection. Genet Resour Crop Evol 59:499–515
Mukamuhirwa F, Tusiime G, Mukankusi C, Gibson P, Edema R (2012) Potential sources of high iron and zinc content in Ugandan bean germplasm. Third RUFORUM Biennial Meeting 24–28 September 2012, Entebbe
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nemli S, Asciogul TK, Kay HB, Kahraman A, Siyok DE, Tanyolac B (2014) Association mapping for five agronomic traits in the common bean (Phaseolus vulgaris L.). J Sci Food Agric. doi:10.1002/jsfa.6664
Nemli S, Asciogul TK, Esiyok DDA, Tanyolac MB (2016) Genome wide association mapping (GWAS) of nutrient elements in seed using genotyping by sequencing (GBS) in common bean (Phaseolus vulgaris L.). Proceeding of plant and animal genome conference XXIV
Okii D, Tukamuhabwa P, Kami J, Namayanja A, Paparu P, Ugen M, Gepts P (2014) The genetic diversity and population structure of common bean (Phaseolus vulgaris L.) germplasm in Uganda. Afr J Biotech 13(29):2935–2949
Pennington JAT, Young B (1990) Iron zinc copper manganese selenium and iodine in foods from the United States total diet study. Food Compos Anal 3:166–184
Perrier X, Jacquemoud-Collet JP (2006) DARwin software, http://darwin.cirad.fr/darwin
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ramasamy RK, Ramasamy S, Bindroo BB, Naik VG (2014) STRUCTURE PLOT: a program for drawing elegant STRUCTURE bar plots in user friendly interface. Springerplus 13(3):431. doi:10.1186/2193-1801-3-431
Salvi S (2007) Conserved non-coding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA 104:11376–11381
Sgarbieri VC, Antunes PL, Almeida LD (1979) Nutrional evaluation of four varieties of dry beans (Phaseolus vulgaris L.). J Food Sci 44:1306–1308
Shehzad T, Iwata H, Okuno K (2009) Genome-wide association mapping of quantitative traits in sorghum (Sorghum bicolor (L.) Moench) by using multiple models. Breed Sci 59:217–227
Shi C, Navabi A, Yu K (2011) Association mapping of common bacterial blight resistance QTL in Ontario bean breeding populations. BMC Plant Biol 11:52
Silva VR, Iachan A (1975) Proteínas de variedadesbrasileiras de feijão (P. vulgaris). I- Quantificação e fracionamento das proteínas. Rev Bras Tecnol 6:133–141
Silva CA, Abreu AFB, Ramalho MAP, Correa DA, Maia LGS (2010) Genetic variability for protein and minerals content in common bean lines (Phaseolus vulgaris L.). Ann Rep Improv Co-op 53:144–145
Sood S, Awasthi CP, Singh N (2003) Biochemical evaluation of promising Rajmash (Phaseolus vulgaris L.) genotypes of Himachal Pradesh, Himachal. J Agric Res 29(1&2):65–69
Talukder ZI, Anderson E, Miklas PN, Blair MW, Osorno J, Dilawari M, Hossain KG (2010) Genetic diversity and selection of genotypes to enhance Zn and Fe content in common bean. Can J Plant Sci 90:4960
Tezoto SS, Sgarbieri VC (1990) Protein nutritive value of a new cultivar of bean (Phaseolus vulgaris L.). J Agr Food Chem 38:1152–1156
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES (2001) Dwarf 8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289
Tryphone GM, Nchimbi-Msolla S (2010) Diversity of common bean (Phaseolus vulgaris L.) genotypes in iron and zinc contents under screen house conditions. Afr J Agric Res 5(8):738–747
Wei XM, Jackson PA, McIntyre CL, Aitken KS, Croft B (2006) Associations between DNA markers and resistance to diseases in sugarcane and effects of population substructure. Theor Appl Genet 114:155–164
Welch RM, Graham RD (2004) Breeding for micronutrients in staple food crops from a human nutrition perspective. J Exp Bot 55:353–364
Welch RM, House WA, Beebe S, Cheng Z (2000) Genetic selection for enhanced bioavailable levels of iron in bean (Phaseolus vulgaris L.) seeds. J Agric Food Chem 48:3576–3580
Wilson LM, Whitt SR, Rocheford TR, Goodman MM, Buckler ES (2004) Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16:2719–2733
Yu K, Park SJ, Poysa V, Gepts P (2000) Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). Heredity 9:429–434
Zhang Y, Sun Y, Ye Y, Karim MR, Xue Y, Yan P, Meng Q, Cui Z, Cakmak I, Zhang F (2012) Zinc biofortification of wheat through fertilizer applications in different locations of China. Field Crops Res 125:1–7
Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Prize AH, Norton GJ, Islam MR, Reynolds A, Mezey J, McClung AM, Bustamante CD, McCouch SR (2011) Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2:467
Zondervan KT, Cardon LR (2004) The complex interplay among factors that influence allelic association. Nat Rev Genet 5:89–100
Acknowledgements
SMZ is grateful to SERB, New Delhi for financial support of this work (Project Sanction Order No. SR/FT/LS-27/2011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13205_2017_928_MOESM1_ESM.ppt
Supplementary material 1 (PPT 238 kb). Mean ± SE of mineral (Fe and Zn) and protein concentrations in seeds of common bean and their correlation with each other (blue bars indicates Fe concentration, red bars indicates Zn concentration and green bars indicates protein concentration). r value represents the correlation
13205_2017_928_MOESM3_ESM.docx
Supplementary material 3 (DOCX 60 kb). Supplementary Table 1: List of SSR marker used in this study. Supplementary Table 2: Variation among Fe, Zn, and protein contents in diverse common bean genotypes. Supplementary Table 3: Distribution of 138 common bean genotypes on the bases of their mean value range of Fe, Zn and Protein. Supplementary Table 4: Assignment of individuals to the sub populations (K) based on probability
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mahajan, R., Zargar, S.M., Salgotra, R.K. et al. Linkage disequilibrium based association mapping of micronutrients in common bean (Phaseolus vulgaris L.): a collection of Jammu & Kashmir, India. 3 Biotech 7, 295 (2017). https://doi.org/10.1007/s13205-017-0928-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-0928-x