Abstract
Acute and chronic pollution of environments with crude oil does not bode well for biota living within the vicinity of polluted environments. This is due to environmental and public health concerns on the negative impact of crude oil pollution on living organisms. Enhancing microbial activities by adding nutrients and other amendments had proved effective in pollutant removal during bioremediation. This study was carried out to determine how microbial group respond during remediation by enhanced natural attenuation (RENA) during a field-scale bioremediation. Crude oil-polluted soil samples were collected (before, during, and after remediation) from a site undergoing remediation by enhanced natural attenuation (RENA) at Ikarama Community, Bayelsa State, Nigeria, and were analyzed for total petroleum hydrocarbon (TPH), polyaromatic hydrocarbon (PAH), and a shift in microbial community. The gas chromatography-flame ionization detector (GC-FID) results showed that the pollutant concentrations (TPH and PAH) reduced by 98 and 85%, respectively, after the remediation. Culturable hydrocarbon utilizing bacteria (CHUB) was highest (8.3 × 104 cfu/g) for sample collected during the remediation studies, whilst sample collected after remediation had low CHUB (6.1 × 104 cfu/g) compared to that collected before remediation (7.7 × 104 cfu/g). Analysis of 16S rRNA of the isolated CHUB showed they belonged to eight bacterial genera namely: Achromobacter, Alcaligenes, Azospirillus, Bacillus, Lysinibacillus, Ochrobactrum, Proteus, and Pusillimonas, with Alcaligenes as the dominant genus. In this study, it was observed that the bacterial community shifted from mixed group (Gram-positive and -negative) before and during the remediation, to only the latter group after the remediation studies. The betaproteobacteria groups were the dominant isolated bacterial phylotype. This study showed that RENA is an effective method of reducing pollutant concentration in crude oil-polluted sites, and could be applied to other polluted sites in the Niger Delta region of Nigeria to mitigate the devastating effects of crude oil pollution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pollution of environments (air, land and water) has been of concern to the society, especially in developing countries witnessing tremendous and rapid industrialization. One of the classes of pollutants that posses an imminent danger is hydrocarbons. In this class of pollutant, the polyaromatics are more environmental and public health concerns due to their carcinogenic, mutagenic, recalcitrant, and other detrimental effects on living organisms (Erdogmus et al. 2015). Pollution with crude oil can occur either by natural or artificial means. Accidental discharge and spillage during crude oil exploration are the major cause of the former, whilst the causes of the latter are vast ranging from emission from exhausts, indiscriminate discharge of used crude product(s) emanating from automobile repair and construction sites into surrounding water bodies and land. Vandalization of oil pipelines and illegal bunkering are the leading cause of anthropological crude oil pollution in developing countries. To reclaim crude oil-polluted sites, attention has been drawn to bioremediation owing to its eco-friendly, non-invasive, and cost-effective features (Adebusoye et al. 2007; Chikere et al. 2011; Silva-Castro et al. 2015; Dados et al. 2015; Nwogu et al. 2015). However, for a successful bioremediation to be achieved, environmental (abiotic) and biotic factors that might hinder the rate of bioremediation are given major considerations. Microorganisms are known to play vital role in bioremediation. Some may degrade pollutants by utilizing them as carbon source, while others may transform or immobilize pollutant, thus reducing the concentration to a less harmful state. With regards to microbial population in a polluted environment, there are two major approaches to improve microbial activities on pollutant utilization as carbon source: bioaugmentation and biostimulation (Azubuike et al. 2016). Despite the limitations of bioaugmentation (Megharaj et al. 2011; Gao et al. 2014; Shahi et al. 2016), it can be used to remediate sites polluted with recalcitrant pollutants (Philp and Atlas 2005). Nevertheless, biostimulation is a more cost-effective and practical approach to bioremediation (Simpanen et al. 2016). Remediation by enhanced natural attenuation (RENA), which involves improving nutrient, aeration, and moisture content is a good example of land farming in situ bioremediation approach, which have recorded many successes towards pollutant removal (Okukoya and Lambert 2015). Thus, different organic and inorganic substrates have been used as biostimulants to expedite bioremediation process during RENA. However, most of the reductions in pollutant concentrations with such biostimulants were obtained during small-scale studies. Lack of access to polluted sites and cost of field-scale bioremediation contribute to difficultly in replicating lab-scale experiment at field-scale level. Further, pollutant inhomogeneity coupled with changes in climate contributes to variability in results obtained during field-scale bioremediation (Garg et al. 2016). This makes it difficult to directly compare the results of lab-scale with that of field-scale bioremediation studies (Simpanen et al. 2016). This study was carried out to determine bacterial community shift and the efficacy of RENA towards pollutant removal during field-scale bioremediation project at Okodia-Rumuekpe, Ikarama Community, Yenagoa, Bayelsa State, Nigeria.
Materials and methods
Site description and sample collection
The soil samples used in this study were collected from a polluted site at Okodia-Rumuekpe, Ikarama Community, Yenagoa, Bayelsa State, Nigeria; a site undergoing field-scale remediation by enhanced natural attenuation (RENA). Accordingly, the observed treatments made to the polluted site were nutrient addition (nitrogen, phosphorus and potassium at ratio of 2:1:1), tilling, irrigation, and intermittent turning (windrow construction and breakdown) to ensure uniform aeration. The global positioning system (GPS) coordinates of the site are: Northing (05° 081 07.3) and Easting (006° 281 07.5). At every sampling interval (3 weeks), samples were collected at depth of 0–15 and 0–30 cm from different points and pooled to obtain composite sample, which were used for further analyses. The polluted site was monitored pre (before), during, and post (after) remediation. The control soil sample (control) was collected 100 m away from the polluted area undergoing RENA. Samples were aseptically transported to Microbiology laboratory and preserved as described (Sojinu et al. 2010). Media used for the analyses were of analytical grade and were obtained from Sigma-Aldrich, United States.
Determination of physicochemical parameters and oil chemistry
Parameters such as total organic carbon (TOC), nitrogen, pH, conductivity, and phosphorus contents were determined using the methods described in APHA (2008). Total hydrocarbon, total petroleum, and aromatic hydrocarbons (TPH and PAH, respectively) contents were determined using gas chromatography-flame ionization detector as described (USEPA 2001; ASTM 2010).
Enumeration of culturable heterotrophic and hydrocarbon utilizing bacteria
The total culturable heterotrophic bacterial (TCHB) counts were determined using plate count agar (PCA) as described by Chikere and Ekwuabu (2014). Soil suspensions were prepared by a tenfold serial dilution of 1 g of soil in 9 mL sterile normal saline (0.85% NaCl w/v in distilled water). An aliquot of 0.1 mL serially diluted sample was transferred to prepared PCA plates, and spread evenly. The inoculated PCA plates were incubated at 37 °C for 24 h. Culturable hydrocarbon utilizing bacteria (CHUB) were enumerated as described (Hamamura et al. 2006; Nwogu et al. 2015) using Bushnell-Haas (mineral salt) agar with Okono medium crude oil as a carbon source. The cultured plates were incubated at 37 °C for 7 days or more, until visible colonies appeared. Following incubation, CHUB isolates were sub-cultured, and preliminary identification was carried out based on their morphological and biochemical characteristics with reference to Holt et al. (1994).
Hydrocarbon degradation screening
Representative hydrocarbon utilizing bacteria isolates were subjected to further screening for oil degradation capabilities. Briefly, a loopful of 18 h old culture of each CHUB was transferred into 9 mL physiological saline and incubated for 4–6 h to achieve 0.5 McFarland standards, which measures a cell concentration 1.0 × 108 cfu/mL, and was subsequently transferred into Bushnell-Haas broth containing 1% (v/v) crude oil. Biodegradation was scored by turbidity, emulsification of oil in Bushnell-Haas broth, and optical density measurement using spectrophotometer after 21-day incubation at 37 °C (Peressutti et al. 2003; Chikere and Ekwuabu 2014).
Microbial community analysis
Bacterial isolates that showed positive for crude oil degradation were transferred to 5 mL tube containing Lauria Bertani (LB) broth and incubated at 37 °C for 24 h. Following this, genomic DNAs were extracted using Zymo Research Bacterial DNA MiniPrep Kit according to manufacturer’s instruction. Extracted DNAs were quantified using a Thermo Scientific Nanodrop (ND) 1000 spectrophotometer and were analyzed by gel electrophoresis. Subsequently, purified extracted DNAs were amplified using a 16S rRNA eubacterial universal primer set, 27F (5′AGAGTTTGATCMTGGCTCAG3′) and 1492R (5′TACGGYTACCTTGTTACGACTT3 (Jiang et al. 2006; da Silva et al. 2013). The total PCR mixture was 25 µL and contained the followings: Zymo Master Mix™ (12.5 µL), forward and reverse primer (0.8 µL each), DNA template (5 µL), and sterile PCR water (5.9 µL). The reaction was carried in a thermal cycler as follows: initial denaturation temperature of 95 °C for 5 min, denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s, extension at 72 °C for 45 s, and final extension at 72 °C for 3 min. Following the thermal cycling reaction, 10 µL of each amplicon was analyzed by electrophoresis using 1% (w/v) agarose gel prepared in Tris–borate–EDTA (TBE) buffer. The gels were visualized using UV Trans-illuminator.
Furthermore, amplicons were sequenced using chain termination method of Sanger with a 3500 ABI genetic analyzer. The sequences generated were visualized using the software Chromaslite followed by base calling. BioEdit was used for sequence editing prior to nucleotides search for similarities at basic local alignment search tool (BLAST-N) of the National Center for Biotechnology Information (NCBI) database.
Statistical analyses
One-way analysis of variance (ANOVA) and multiple comparison tests (posthoc) were used to determine level of significance of the data generated in this study. The analysis was carried out using Statistical Package for Social Science (SPSS) Version 20.0. Descriptive statistics such as mean, standard deviation, and standard error of means were carried out. Further, post hoc was performed at 5% significant difference level (P = 0.05) in other to access significant changes in the measured variables.
Results
Determination of physicochemical parameters and oil chemistry
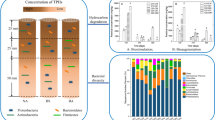
Crude oil-polluted soil samples were collected from Ikarama, a site undergoing remediation by enhanced natural attenuation (RENA) and were analyzed for reduction in pollutant concentration, and corresponding shift in microbial population pre, during, and post remediation. The baseline analyses show that the site (Supplementary Information, Fig. 1S) was indeed polluted with heavy fractions of hydrocarbons, with initial total petroleum hydrocarbons (TPH) and polyaromtic hydrocarbons (PAHs) contents of 16,618 and 1017 mg/kg, respectively. The control soil sample had TPHs and PAHs contents of 277 and 7.30 mg/kg, respectively. It was clear from the baseline analyses that total phosphorus content was higher than nitrogen content for both polluted and control sites. Nitrogen content was higher for polluted site compared to control site, whilst phosphorus content was higher for unpolluted site as compared to polluted site (Table 1). Composite soil sample collected after the remediation showed drastic reduction in the concentrations of TPHs and PAHs, 98 and 85%, respectively (Fig. 1). Interestingly, 96% of the reduction was obtained for TPHs, and 84.7% for PAHs during the remediation. Three-ringed aromatic compounds were the dominant PAHs detected (46.76%), followed by 4-ringed (32.11%), and 5-ringed (12.53%). Two-ringed aromatic hydrocaron, naphthalene, constituted the lowest proportion (0.19%) of detected PAHs in the polluted sample before RENA was initiated. The remaining percentages: 8.2 and 0.2% were for 7-ringed Indeno [1,2,3-c,d] perylene and 6-ringed Benzo[ghi] perylene PAHs, respectively. Furthermore, amongst the PAHs detected in the polluted samples before remediation, dibenz[g,h,i]anthracene, ideno[1,2,3-c,d]perylene, and benzo[g,h,i]perylene were not detected during, and after the remediation. Benz[a]anthracene and chrysene were reduced by 76.3 and 79.5%, respectively, during the remediation. However, they were not detected in the sample taken after the RENA (Fig. 2). In the pre-sample for PAHs, fluorene had the highest concentration, 133.9 mg/kg, followed by anthracene, 132.5 mg/kg, and the lowest/detectable concentration, 1.9 mk/kg was recorded for naphthalene.
Enumeration of culturable heterotrophic and hydrocarbon utilizing bacteria
The total culturable heterotrophic bacteria (TCHB) and culturable hydrocarbon utilizing bacteria (CHUB) were obtained pre, during, and after the remediation. The crude oil-polluted soil before RENA treatment had the highest TCHB, with a count of 8.6 × 104 cfu/g followed by that of control sample, 8.4 × 104 cfu/g whereas the lowest count, 5.4 × 104 cfu/g was obtained from the sample collected during the RENA. In contrast, CHUB for polluted sample collected during the remediation recorded highest microbial count, 8.3 × 104 cfu/g. As expected, the lowest CHUB count, 3.3 × 104 cfu/g was recorded for the control sample (Fig. 3). There was no significant difference (P = 0.07) in TCHB across all the different samples, both polluted and control. Nevertheless, there was significant difference (P < 0.05) in CHUB obtained in this study.
Changes in microbial counts (log10 cfu/g) for control soil sample (UP), and soil samples collected from polluted sites before (BR), during (DR), and after (AR) remediation by enhanced natural attenuation (RENA). Total culturable heterotrophic bacteria (TCHB) and culturable hydrocarbon utilizing bacteria (CHUB). Bars represent standard errors, n = 2
Hydrocarbon degradation screening
The hydrocarbon utilizing bacteria isolated by vapor phase transfer technique were subjected to hydrocarbon degradation screening in a mineral salt medium containing 1% (v/v) Okono medium crude oil. Each isolate was allowed to grow for 21 days, and turbidity was measured using a spectrophotometer at every 7-day interval. Out of the thirteen isolates from control sample, two showed crude oil degradability capability, with gradual increase in turbidity from day 0 through to day 21 (Fig. 4a). Amongst the 11 from polluted sample before remediation, 5 showed positive for crude oil degradation, whilst those from sample during remediation were seven positive isolates (Fig. 4b, c). In addition, seven out of ten isolates from sample after remediation were positive for crude oil degradation (Fig. 4d). Interestingly, only two of the positive isolates (BR1 and DR9) had optical densities <0.8 at 600 nm. Meanwhile, the highest optical density (1.60) was recorded for isolate AR10 from sample after remediation. Generally, isolates from sample after remediation had higher optical densities compared to others from the control, pre, and during RENA. Furthermore, one-way analysis of variance (ANOVA) and multiple comparison tests (posthoc) showed significant difference (P < 0.05) in hydrocarbon utilization (based on optical density readings) during day 7, 14, and 21 of the degradation screening for all the samples. Nevertheless, at day-0, there was no significant difference (P = 0.426) in optical density readings for all the isolates obtained from all the samples sources.
Hydrocarbon degradation screening of culturable hydrocarbon utilizing bacterial (CHUB) isolates obtained by vapor phase transfer method. a Isolates from control (UP) soil sample. b Isolates before (BR); c isolates during (DR); d isolates after (AR) remediation by enhanced natural attenuation (RENA). The number after each two-letter code for each sample source, represents isolate code. The degradation was monitored for 21 days at interval of 7 days. Bars represent standard errors, n = 2
Microbial community analysis
The 21 bacterial isolates which utilized crude oil as sole carbon source as evidenced by the hydrocarbon degradation screening were used for further studies. DNA extraction was successfully carried out on each isolates (Supplementary Information, Figure S2) and the extracted DNAs were amplified using a set of universal primer. Sequence amplification was successful for 19 of the isolates except BR1 and DR9, which were not successfully amplified. Therefore, the 19 isolates were further subjected to sequencing with Sanger method. All of the isolates were successfully sequenced and each sequence was matched with the closest sequence in the National Centre for Biotechnology Information (NCBI) GeneBank using basic local alignment search tool (BLAST). The identity of the isolated bacterial species covered 8 genera: Achromobacter, Alcaligenes, Azospirillum, Bacillus, Lysinibacillus, Ochrobactrum, Proteus, and Pusillimonas (Table 2). Amongst the genera, two (Bacillus and Lysinibacillus) were Gram-positive; others were Gram-negative. Both Gram-positive and betaproteobacteria were isolated from the control sample. The soil sample collected pre (before) RENA was initiated, had 50% Gram-positive and 50% γ-proteobacteria. On the other hand, sample during remediation had alpha, beta, and Gram-positive bacterial population. At the end of the remediation, it became apparent that only Gram-negative bacteria were isolated amongst which 86% were betaproteobacteria, and 14% were gammaproteobacteria. In general, it was observed that the bacterial population in this study shifted from both Gram-positive and negative groups during the remediation, to only the latter group after the RENA. More so, very few betaproteobacteria were isolated during the remediation, but this group dominated after the remediation when pollutant concentration appeared to have been reduced to an innocuous state.
Discussion
In this study, the effect of remediation by enhanced natural attenuation (RENA) on autochthonous bacterial population was monitored pre (before), during, and after RENA process. The processes in the RENA treatment were: nutrient addition (inorganic fertilizer), tilling, irrigation, and periodic turning to ensure uniform aeration. The polluted soil sample used in this study was obtained from a crude oil-impacted site undergoing RENA at Ikarama, Bayelsa State, Nigeria. The high phosphorus contents for both polluted and control sites compared to nitrogen was attributed to the nature of the sites. The sites have not witnessed agricultural activities in the past few decades, as it is a crude oil pipeline trajectory, which is prohibited of any activity. However, in the polluted site, the difference was not as much as it was for the control site (Table 1). This might be as a result of gradual utilization of phosphorus by resident microbes present in the polluted site, for degradation of the pollutant. This observation was in contrast to two-times phosphorus difference in the control site, which was attributable to less degradation and microbial activities. Phosphorus has been reported as essential element for maintaining vitality in living organisms owing to its energy storage and processing of genetic information (Elser 2012). The reduction in both total petroleum hydrocarbons (TPHs) and polyaromatic hydrocarbons (PAHs) concentrations were attributed to the RENA treatments. Addition of inorganic fertilizer as a biostimulant has been reported to increase microbial activities in polluted environments and resulted in increase pollutant removal rate (Belochini et al. 2010; Teng et al. 2010; Kauppi et al. 2011; Agarry and Ogunleye 2012; Silva-Castro et al. 2013, 2015). It has been reported that aeration by tillage and appropriate moisture content are sufficient to stimulate microbial activities during bioremediation in some soils (Silva-Castro et al. 2015). This could account for the enhanced pollutant removal, TPHs (96%) and PAHs (84.7%) observed for sample collected during this study as opposed to that collected before the study commenced. The lowest PAH detection recorded for naphthalene was as a result of its low molecular weight (LWM) nature, which contributed to volatilization in the environment shortly after pollution. In contrast, the high molecular weight (HMW) nature, high hydrophobicity, and low transport potential of 4–5 ringed PAH members (Kuppusamy et al. 2016) could have contributed to persistence of these PAHs group during and after the remediation as observed in this RENA studies. Nevertheless, the inability to detect some 4-ringed PAHs (benz[a]anthracene and chrysene) after the remediation could be as a result of complete degradation and mineralization given the rapid reductions in concentrations (76.3 and 79.5%, respectively) of these constituents within few weeks of the RENA. Similarly, the failure to detect 5–7 ringed PAHs during the remediation was also attributable to degradation and mineralization. Thus, it was possible that the RENA treatment encouraged co-metabolism within the native PAH degraders present in the polluted site. A functional metagenomic study by Zafra et al. (2016) reported a faster PAH removal ascribable to microbial co-metabolism. Notwithstanding, pollutant non-bioavailability due to less solubility and decreased mass transfer especially for PAHs, which increases with increased molecular weight (Fuentes et al. 2014) could also be accountable for the lack of detection of heavier PAH fractions during and after the remediation. Furthermore, it was plausible that some of the heavier PAHs were transformed and were not detected in their original forms.
The high total culturable heterotrophic bacteria (TCHB) obtained for control sample was due to low concentration of pollutant (TPHs and PAHs), which in turn encouraged the proliferation of heterotrophs, and supported their growth and utilization of other organic carbons present in the control soil as source of energy. On the other hand, the low concentration of pollutant did not favor the growth of culturable hydrocarbon utilizing bacteria (CHUB) as a result, the control sample recorded the lowest CHUB count. The highest CHUB recorded for sample collected during the remediation was attributed to biostimulation of the polluted site with nutrients and other treatments, which favored, and stimulated the activities of hydrocarbonoclastic bacteria resulting in rapid pollutant degradation; similar observation has been reported (Onuoha 2013). The reduced CHUB count obtained after the remediation was ascribed to reduction in pollutant concentration and nutrient depletion (Adebusoye et al. 2007; Shabir et al. 2008; Nwogu et al. 2015). These two factors tended to encourage the re-establishment of TCHB in the site after the treatment as a result there was increased TCHB compared to that obtained during the remediation. Natural selection, biostimulation of indigenous oleophilic microbes, biogenic synthesis, natural seepage of hydrocarbon could account for the presence of CHUB even in the control soil ecosystems (Bissett et al. 2013; Macauley and Rees 2014; Liang et al. 2016).
It was observed that not all the isolated CHUB were able to actively degrade crude oil during the degradation screening, despite the fact that same crude oil was used for their preliminary isolation. This observation might be due to the vapor phase transfer method used during the preliminary isolation. Thus, it was likely that impurities in agar, and the ability of agar to absorb nutrient resulted in the growth and isolation of non-crude oil degraders as CHUB (Chikere et al. 2011). In addition, not all hydrocarbon degraders, especially short and long chain alkane degraders can utilize hydrocarbons in the volatile state of vapor phase transfer method (Wang and Shao 2012, 2013). Therefore, degradation screening is an effective way of thinning out the numbers of non-crude oil degraders, which might have been regarded as hydrocarbon degraders due to biases originating from vapor phase transfer method (Hamamura et al. 2006). Further, given that single CHUB isolate was subjected to the degradability screening with crude oil containing different hydrocarbon fractions as carbon source, it was likely that the CHUB, which did not show active crude oil degradation, were unable to utilize the hydrocarbons as carbon source when present as a mixture of different fractions. Apparently, it does not mean these particular CHUB do not posses degradation potentials. It has been reported that CHUB, which originally were able to degrade some crude oil fractions (petrol and engine oil) on separate occasions were unable to do so when both fractions were mixed (Shankar et al. 2014). More so, it has been reported that single bacteria species tended to exhibit limited hydrocarbon degradation compared to consortia, owing to wide system of degradative enzyme in the latter (Hassanshahian et al. 2014; Nkem et al. 2016). It is not surprising that isolates from control sample were positive for hydrocarbon degradation. This was attributable to the residual fractions of both TPHs and PAHs detected in the sample, which continued to serve as carbon sources for the hydrocarbonoclastic bacteria (Bacillus cereus and Pusillimonas sp.) isolated from the unpolluted site. Similarly, Peressutti et al. (2003) isolated Bacillus brevis from control soil with 0.7% (w/w) hydrocarbon concentration. The high optical densities, and numbers of crude oil degrading bacteria obtained from samples during and after remediation in comparison to those from sample before remediation were also attributed to biostimulation by the RENA process. In addition, the innate ability of most of the identified positive hydrocarbon degrading genera (>70% Alcaligenes spp.) isolated from sample after remediation contributed to their high optical densities during the degradation screening. Members of Alcaligenes have been reported as versatile hydrocarbon degraders, which posse alkB gene that code for enzyme involved in hydrocarbon degradation (Peressutti et al. 2003; Martínez-Pascual et al. 2015).
The predominance of Gram-negative bacteria, especially the betaproteobacteria phylotype after the remediation as observed in this study has also been reported (Bacosa et al. 2010; Martínez-Pascual et al. 2015). These observations were based on the fact that most of the functionally diverse bacterial group falls with the proteobacteria phylum (Gupta 2000). Similarly, Kuppusamy et al. (2016) in a pyrosequencing analysis of soils chronically polluted with PAHs and heavy metals observed that proteobacteria dominated amongst the operational phylogenic units (OPU). Further, Tauler et al. (2016) also reported 80% proteobacteria phylum dominance during PAH-remediation studies in an enriched soil consortium. Nutrient addition during the RENA process stimulated these bacterial groups (Gram-negative), and boost their competitive advantage (Shahi et al. 2016) and metabolic capabilities (Peresutti et al. 2003) over other bacterial communities in soil polluted with hydrocarbons. This resulted in a shift from mixed population of Gram-positive and negative bacteria to only Gram-negative bacteria at the end of the RENA treatment. In this study, both Gram-positive and -negative bacteria were isolated at same frequency (50%) from sample collected before and during remediation. Recently, Bastida et al. (2016) reported that the resistance mechanism of Gram-negative bacteria to hydrocarbon was attributed to membrane modification (variations in membrane fluidity), whilst that of Gram-positive was likely due to changes in biomass. Similarly, Lazaroaie (2010) attributed the ability of Gram-negative bacteria group to withstand pollutant in soil, especially PAH, to cell wall composition of this bacterial group. The isolation of alphaproteobacteria particularly Ochobactrum sp. only during the remediation was ascribed to the PAH degrading ability and bioemulsifier production capability of the genera (Calvo et al. 2008; Wu et al. 2009; Arulazhagan and Vasudevan 2011), which likely contributed to enhanced pollutant solubility and degradation during the RENA process. In general, other factors such as co-metabolism, syntrophism, mutualism, and antagonism might also be responsible for microbial community changes observed in this study, as these factors have all been reported by several researchers (Morris et al. 2013; Bissett et al. 2013; Dolfing 2013; Luo et al. 2014; Tiantian et al. 2015) to induce shifts in microbial community structure following anthropogenic disturbances.
Conclusion
This study showed that remediation by enhanced natural attenuation (RENA) is an effective means of reducing pollutant (hydrocarbon) concentration especially in the Niger Delta region of Nigeria, which is facing acute and chronic crude oil pollution of environments (soil and water bodies). In this study, it was observed that microbial population shifted from Gram-negative (alphaproteobacteria, and betaproteobacteria) and Gram-positive bacteria during the remediation process to only Gram-negative phylotype (gammaproteobacteria and betaproteobacteria) after the remediation studies.
References
Adebusoye SA, Ilori MO, Amund OO, Teniola OD, Olatope SO (2007) Microbial degradation of petroleum hydrocarbons in a polluted tropical stream. World J Microbio Biotechnol 23:1149–1159. doi:10.1007/s11274-007-9345-3
Agarry ES, Ogunleye OO (2012) Box–Behnken design application to study enhanced bioremediation of soil artificially contaminated with spent engine oil using biostimulation strategy. Int J Energy Environ Eng 3:31. doi:10.1186/2251-6832-3-31
APHA (2008) Standard methods for the examination of water and wastewater 21st, edn. American Publich Health Association, Washington, DC
ASTM (2010) Test method for oil and grease and petroleum hydrocarbons in water, D3921 (11.02). In: Annual book of ASTM standrads. American Society for Testing and Materials, Philadephia, PA, USA
Arulazhagan P, Vasudevan N (2011) Biodegradation of polycyclic aromatic hydrocarbons by a halotolerant bacterial strain Ochrobactrum sp. VA1. Mar Pollut Bull 62:388–394. doi:10.1016/j.marpolbul.2010.09.020
Azubuike CC, Chikere CB, Okpokwasili GC (2016) Bioremediation techniques–classification based on site of application: principles, advantages, limitations and prospects. World J Microbiol Biotechnol 32:1–18. doi:10.1007/s11274-016-2137-x
Bacosa H, Suto K, Inoue C (2010) Preferential degradation of aromatic hydrocarbons in kerosene by a microbial consortium. Int Biodeterior Biodegrad 64:702–710. doi:10.1016/j.ibiod.2010.03.008
Bastida F, Jehmlich N, Lima K, Morris BEL, Richnow HH, Hernández T, von Bergen M, García C (2016) The ecological and physiological responses of the microbial community from a semiarid soil to hydrocarbon contamination and its bioremediation using compost amendment. J Proteo 135:162–169. doi:10.1016/j.jprot.2015.07.023
Beolchini F, Rocchetti L, Regoli F, Dell’Anno A (2010) Bioremediation of marine sediments contaminated by hydrocarbons: experimental analysis and kinetic modeling. J Hazard Mater 182:403–407. doi:10.1016/j.jhazmat.2010.06.047
Bissett A, Brown MV, Siciliano SD, Thrall PH (2013) Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecol Lett 16:128–139. doi:10.1111/ele.12109
Calvo C, Silva-Castro GA, Uad I, García FC, Laguna J, González-López J (2008) Efficiency of the EPS emulsifier produced by Ochrobactrum anthropi in different hydrocarbon bioremediation assays. J Ind Microbiol Biotechnol 35:1493–1501. doi:10.1007/s10295-008-0451-5
Chikere CB, Ekwuabu CB (2014) Culture-dependent characterization of hydrocarbon utilizing bacteria in select crude oil-impacted sites in Bodo, Ogoniland, Nigeria. Afr J Environ Sci Technol 8:401–406. doi:10.5897/AJEST2014.1707
Chikere CB, Okpokwasili GC, Chikere BO (2011) Monitoring of microbial hydrocarbon remediation in the soil. 3 Biotech 1:117–138. doi:10.1007/s13205-011-0014-8
da Silva MAC, Cavalett A, Spinner A, Rosa DC, Jasper RB, Quecine MC, Bonatelli ML, Pizzirani-Kleiner A, Corção G, Lima AOS (2013) Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. SpringerPlus 2:127. doi:10.1186/2193-1801-2-127
Dados A, Omirou M, Demetriou K, Papastephanou C, Ioannides IM (2015) Rapid remediation of soil heavily contaminated with hydrocarbons: a comparison of different approaches. Ann Microbiol 65:241–251. doi:10.1007/s13213-014-0856-5
Dolfing J (2013) Syntrophic degradation of propionate via butyrate: a novel window of opportunity under methanogenic conditions. J Appl Environ Microbio 79:4515–4516. doi:10.1128/AEM.00111-13
Elser JJ (2012) Phosphorus: a limiting nutrient for humanity? Curr Opin Biotechnol 23:833–838. doi:10.1016/j.copbio.2012.03.001
Erdogmus SF, Korcan SE, Konuk M, Guven K, Mutlu MB (2015) Aromatic hydrocarbon utilization ability of Chromohalobacter sp. Ekologi. 24:10–16. doi:10.5053/ekologi.2015.942
Fuentes S, Méndez V, Auguila P, Seeger M (2014) Bioremediation of petroleum hydrocarbons: catabolic genes, microbial communities, and applications. Appl Microbiol Biotechnol 98:4781–4794. doi:10.1007/s00253-014-5684-9
Gao Y, Guo S, Wang J, Li D, Wang H, Zeng D-H (2014) Effects of different remediation treatments on crude oil contaminated saline soil. Chemospere 117:486–493. doi:10.1016/j.chemosphere.2014.08.070
Garg N, Lata P, Jit S, Sangwan N, Singh AK, Dwivedi V, Niharika N, Kaur J, Saxena A, Dua A, Nayyar N, Kohli P, Geueke B, Kunz P, Rentsch D, Holliger C, Kohler H-PE, Lal R (2016) Laboratory and field scale bioremediation of hexachlorocyclohexane (HCH) contaminated soils by means of bioaugmentation and biostimulation. Biodegr 27:179–193. doi:10.1007/s10532-016-9765-6
Gupta RS (2000) The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev 24:367–402. doi:10.1111/j.1574-6976.2000.tb00547.x
Hamamura N, Olson SH, Ward DM, Inskeep WP (2006) Microbial population dynamics associated with crude oil biodegradation in diverse soils. Appl Environ Microbiol 72:6316–6324. doi:10.1128/AEM.01015-06
Hassanshahian M, Emtiazi G, Caruso G, Cappello S (2014) Bioremediation (bioaugmentation/biostimulation) trials of oil polluted seawater: a mesocosm simulationstudy. Marine Environ Res 95:28–38
Holt JG, Krieg NR, Sneath HA, Staley JJ, Williams ST (1994) Bergey’s manual of determinative bacteriology. Williams and Wilkins, Philadelphia
Jiang H, Dong H, Zhang G, Yu B, Chapman LR, Fields MW (2006) Microbial diversity in water and sediment of Lake Chaka, an Athalassohaline Lake in Northwestern China. Appl Environ Microbiol 72:3832–3845. doi:10.1128/AEM.02869-05
Kauppi S, Sinkkonen A, Romantschuk M (2011) Enhancing bioremediation of diesel-fuel-contaminated soil in a boreal climate: comparison of biostimulation and bioaugmentation. Inter Biodeter Biodegrad 65:359–368. doi:10.1016/j.ibiod.2010.10.011
Kuppusamy S, Thavamani P, Megharaj M, Venkateswarlu K, Lee YB, Naidu R (2016) Pyrosequencing analysis of bacterial diversity in soils contaminated long-term with PAHs and heavy metals: implications to bioremediation. J Hazard Mater 300:169–179. doi:10.1016/j.jhazmat.2016.05.066
Lazaroaie MM (2010) Multiple responses of gram-positive and gram-negative bacteria to mixture of hydrocarbons. Braz J Microbiol 4:1517–8382. doi:10.1590/S151783822010000300016
Liang Y, Zhao H, Deng Y, Zhou J, Li G, Sun B (2016) Long-term oil contamination alters the molecular ecological networks of soil microbial functional genes. Front Microbiol 7:113. doi:10.3389/fmicb.2016.00060
Luo W, Zhu X, Chen W, Duan Z, Wang L, Zhou Y (2014) Mechanisms and strategies of microbial cometabolism in the degradation of organic compounds-chlorinated ethylene as the model. J Int Assoc Water Pollu Res 69:1971–1983. doi:10.2166/wst.2014.108
MaCauley BM, Rees D (2014) Bioremediation of oil spills: a review of challenges for researcher advancement. Ann Environ Sci 8:9–37
Martínez-Pascual E, Grotenhins T, Solanas AM, Viñas M (2015) Coupling chemical oxidation and biostimulation: effects on the natural attenuation capacity and resilience of the native microbial community in alkylbenzene-polluted soil. J Hazard Mater 300:135–143. doi:10.1016/j.jhazmat.2015.06.061
Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R (2011) Bioremediation approaches for organic pollutants: a critical perspective. Environ Int 37:1362–1375. doi:10.1016/j.envint.2011.06.003
Morris BE, Henneberger R, Huber H, Moissi-Eichinger C (2013) Microbial syntrophic: interaction for the common good. FEMS Microbiol Rev 37:384–406. doi:10.1111/1574-6976.12019
Nkem BM, Halimoon N, Yusoff FM, Johari WLW, Zakaria MP, Medipally SR, Kannan N (2016) Isolation, identification and diesel-oil biodegradation capacities of indigenous hydrocarbon-degrading strains of Cellulosimicrobium cellulans and Acinetobacter baumannii from tarball at Terengganu beach, Malaysia. Marine Pollut Bull 107:261–268. doi:10.1016/j.marpolbul.2016.03.060
Nwogu TP, Azubuike CC, Ogubgue CJ (2015) Enhanced bioremediation of soil artificially contaminated with petroleum hydrocarbons after amendment with Capra aegagrus hircus (Goat) manure. Biotechnol Res Int. doi:10.1155/2015/657349
Okukoya J, Lambert R (2015) Remediation by enhanced natural attenuation (RENA): a beneficial strategy for polyaromatic hydrocarbon degradation and agrifood production. Proc World Cong New Tech 183:1–11
Onuoha SC (2013) Stimulated biodegradation of spent lubricating motor oil in soil amended with animal droppings. J Nat Sci Res 3:106–116
Peressutti SR, Alvarez HM, Pucci OH (2003) Dynamics of hydrocarbon-degrading bacteriocenosis of an experimental oil pollution in Patagonian soil. Int Biodeter Biodegra 52:21–30. doi:10.1016/S0964-8305(02)00102-6
Philp JC, Atlas RM (2005) Bioremediation of contaminated soils and aquifers. In: Atlas RM, Philp JC (eds) Bioremediation: applied microbial solutions for real-world environmental cleanup. American Society for Microbiology (ASM), Washington, pp 139–236
Shabir GM, Afzal M, Anwar F, Tahseen R, Khalid ZM (2008) Biodegradation of kerosene in soil by a mixed bacterial culture under different nutrient conditions. Int Biodeterior Biodegrad 61:161–166. doi:10.1016/j.ibiod.2007.06.003
Shahi A, Aydin S, Ince B, Ince O (2016) Evaluation of microbial population and functional genes during the bioremediation of petroleum-contaminated soil as an effective monitoring approach. Ecotoxicol Environ Safety 125:153–160. doi:10.1016/j.ecoenv.2015.11.029
Shankar S, Kansrajh C, Dinesh MG, Satyan RS, Kiruthika S, Tharanipriya A (2014) Application of indigenous microbial consortia in bioremediation of oil-contaminated soils. Int J Environ Sci Technol 11:367–376. doi:10.1007/s13762-013-0366
Silva-Castro GA, Rodelas B, Perucha C, Laguna J, Gonzalez-Lopez J, Calvo C (2013) Bioremediation of diesel-polluted soil using biostimulation as post-treatment after oxidation with fenton-like reagents: assays in a pilot plant. Sci Total Environ 15(445–446):347–355. doi:10.1016/j.scitotenv.2012.12.081
Silva-Castro GA, Uad I, Rodríguez-Calvo A, González- López J, Calvo C (2015) Response of autochthonous microbiota of diesel polluted soils to land-farming treatments. Environ Res 137:49–58. doi:10.1016/j.envres2014.11.009
Simpanen S, Mäkelä R, Mikola J, Silvennoinen H, Romantschuk M (2016) Bioremediation of creosote contaminated soil in both laboratory and field scale: investigating the ability of methyl-b-cyclodextrin to enhance biostimulation. Int Biodeter Biodegr 106:117–126. doi:10.1016/j.ibiod.2015.10.013
Sojinu OSS, Wang J-Z, Sonibare OO, Zeng EY (2010) Polycyclic aromatic hydrocarbons in sediments and soils from oil exploration areas of the Niger Delta, Nigeria. J Hazard Mater 174:641–647. doi:10.1016/j.jhazmat.2009.09.099
Tauler M, Vila J, Nieto JM, Grifoll M (2016) Key high molecular weight PAH-degrading bacteria in a soil consortium enriched using a sand-in-liquid microcosm system. Appl Microbiol Biotechnol 100:3321–3336. doi:10.1007/s00253-015-7195-8
Teng Y, Luo Y, Ping L, Zou D, Li Z, Christie P (2010) Effects of soil amendment with different carbon sources and other factors on the bioremediation of an aged PAH-contaminated soil. Biodegrad 21:167–178. doi:10.1007/s10532-009-9291-x
Tiantian S, Yongrui P, Mutai B, Nana X, Jinren L (2015) Biodegradation of different petroleum hydrocarbons by free and immobilzed microbial consortia. J Environ Sci Pro Impacts 17:2022–2033. doi:10.1039/C5EMoo318k
United States Environmental Protection Agency, USEPA (2001). National risk management research laboratory Cincinnati OH 45268
Wang W, Shao Z (2012) Diversity of flavin-binding monoooxygenase genes (almA) in marine bacteria capable of degradation of long-chain alkanes. FEMS Microbiol Eco 80:523–533. doi:10.1111/j.1574-6941.2012.01322.x
Wang W, Shao Z (2013) Enzymes and genes involved in aerobic alkane degradation. Front Microbiol 4:1–7. doi:10.3389/fmicb.2013.00089
Wu Y, He T, Zhong M, Zhang Y, Li E, Huang T, Hu Z (2009) Isolation of marine benzo[a]pyrene-degrading Ochrobactrum sp. BAP5 and proteins characterization. J Environ Sci 21:1446–1451. doi:10.1016/S1001-0742(08)62438-9
Zafra G, Taylor TD, Absalón AE, Cortées-Espinosa DV (2016) Comparative metagenomic analysis of PAH degradation in soil by a mixed microbial consortium. J Hazard Mater 318:702–710. doi:10.1016/j.jhazmat.2016.07.060
Acknowledgements
This research received no funding from any agency. The authors are grateful to the natives of Okodia-Rumuekpe, Ikarama Community, Yenagoa, Bayelsa State, for peaceful access to the crude oil-polluted site, where samples for this study were collected.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest in the course of this publication.
Additional information
An erratum to this article is available at http://dx.doi.org/10.1007/s13205-017-0867-6.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chikere, C.B., Azubuike, C.C. & Fubara, E.M. Shift in microbial group during remediation by enhanced natural attenuation (RENA) of a crude oil-impacted soil: a case study of Ikarama Community, Bayelsa, Nigeria. 3 Biotech 7, 152 (2017). https://doi.org/10.1007/s13205-017-0782-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-0782-x