Abstract
Viruses are serious threat to chilli crop production worldwide. Resistance screening against several viruses resulted in identifying a multiple virus resistant genotype ‘IHR 2451’. Degenerate primers based on the conserved regions between P-Loop and GLPL of Resistance genes (R-genes) were used to amplify nucleotide binding sites (NBS)—encoding regions from genotype ‘IHR 2451’. Alignment of deduced amino acid sequences and phylogenetic analyses of isolated sequences distinguished into two groups representing toll interleukin-1 receptor (TIR) and non-TIR, and different families within the group confirming the hypotheses that dicots have both the types of NBS-LRR genes. The alignment of deduced amino acid sequences revealed conservation of subdomains P-loop, RNBS-A, kinase2, RNBS-B, and GLPL. The distinctive five RGAs showing specific conserved motifs were subjected to BLASTp and indicated high homology at deduced amino acid level with R genes identified such as Pvr9 gene for potyvirus resistance, putative late blight resistance protein homolog R1B-23 and other disease resistance genes suggesting high correlation with resistance to different pathogens. These pepper RGAs could be regarded as candidate sequences of resistant genes for marker development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chilli (Capsicum annuum L.) is an indispensable commodity in every Indian cuisine due to its spicy taste, appealing colour and flavour of fruits. The viruses viz., Cucumber mosaic cucumovirus (CMV), Chilli veinal mottle potyvirus (ChiVMV), Chilli leaf curl virus (ChiLCV) and Groundnut bud necrosis virus (GBNV) are very problematic and pose serious threat to chilli crop production as the vectors are surging due to their wide adaptability to climate change. In recent years, farmers are not willing for chilli cultivation in India, mainly due to the severe incidence of viruses. Most of the viruses systemically infect the host, after invading to cells they express their proteins replicate and move from one cell to other (Pallas and Garcia 2011). To counteract, plants also have evolved defence mechanism that are mediated by resistant genes (R-genes) (Kang et al. 2005). Most of the R genes encode proteins with nucleotide binding site (NBS) and leucine rich repeat (LRR) region (Ellis et al. 2000).
Resistant gene analogues (RGAs) are potential genomic regions that contain structural motifs, such as the NBS and LRR regions. Through transposon tagging using Mu transposon, Hm1 gene, a plant R-gene have been cloned for the first time in maize (Johal and Briggs 1992). Later several R-genes such as N, M, RPF, Gpa2 and others have been cloned in different crops like eggplant (Reddy et al. 2015), grapevine (Donald et al. 2002), citrus (Deng et al. 2000) and other species (Sharma et al. 2009). The comparison of the sequences of these R-genes has shown that majority possess conserved amino acid motifs consisting of a NBS domain and a hydrophobic domain (HD) with a consensus amino acid sequence, Gly-Leu-Pro-Leu (GLPL), downstream to NBS (Reddy et al. 2015). Molecular markers facilitate and accelerate the disease resistance breeding programme by reducing the frequency of phenotyping in the process of selection across the generations. There are several molecular marker systems and one of them having more relevance in disease resistance is resistant gene analog polymorphism marker technique (Chen et al. 1998). These RGAP markers were used to identify linked markers for different pathogen resistances in different crops (Yi et al. 2013; Basak et al. 2004; Mutlu et al. 2008).
The conserved motifs within these NBS domains make it possible to use degenerate primers and PCR to isolate RGAs (Kanazin et al. 1996; Leister et al. 1998). These RGAs have the potential application in development of closely linked markers or functional resistant gene markers for marker assisted breeding (Hammond-Kosack and Jones 1997). Leister et al. (1996) developed a PCR based method to easily isolate R-gene analogues (RGAs), from a wide variety of plant species, in which they used degenerate primers that amplify between the kinase 1a motif of the NB-ARC domain and the GLPL motif. RGAs have been successfully isolated using the PCR based approach from a wide range of plant species (Sharma et al. 2009), Cucumis (Wan et al. 2010), hot pepper (Wan et al. 2012) and wild eggplant (Zhuang et al. 2012; Reddy et al. 2015). Cloning of resistant genes and development of molecular markers will be highly useful in marker assisted breeding programs.
The main objective of the study was to isolate RGAs in multiple virus resistant line of hot pepper ‘IHR 2451’ using degenerate primers designed to amplify conserved NBS and LRR regions of RGAs. These isolated RGAs would be helpful in developing the PCR based markers linked to disease resistance in peppers and also for cloning of other crop R-genes in the future.
Materials and methods
Plant material and DNA isolation
The existence of ‘Jackpot’ genotypes that serve as a source of resistance for a number of different diseases (Grube et al. 2000a) will be very much useful and can be exploited by breeders. In the present study, one such jackpot genotype viz., IHR 2451 has been identified as a source of resistance to Cucumber mosaic cucumovirus (CMV) and Chilli veinal mottle potyvirus (ChiVMV) (Naresh et al. 2016), was also reported to be resistant to TMV (Tobacco Mosaic Virus) and other Leaf curl viruses (Singh and Thakur 1979). So this ‘Jackpot’ genotype ‘IHR 2451’ was used to isolate RGAs using PCR based approach. Leaf sample from the 15 days old seedling of ‘IHR2451’ was collected and DNA was isolated by cetyl tri methyl ammonium bromide (CTAB) method (Doyle and Doyle. 1990) with minor modifications. The integrity of isolated purified DNA was determined by gel electrophoresis and DNA was quantified by using Nanodrop (Thermo scientific) and diluted accordingly to final concentration of 40 ng/μl for PCR analysis.
Degenerate primers and PCR amplification
Two pairs of degenerate primers (Table 1) were used to isolate the RGA sequences by amplifying the region between the two conserved domains i.e. P-loop and GLPL in the plant R-genes. These two sets of degenerate primers amplify approximately 500 bp DNA fragments. PCR reaction was performed in final volume of 25 μl, containing 50 ng template DNA, 1× PCR buffer, 2.5 mmol/l MgCl2, 0.5 mmol/l dNTPs, 0.25 mmol/l of each primer and 1.5 units of Taq DNA polymerase (Bangalore, Genei). The reaction was performed in eppendorf thermal cycler and reaction cycle program was with initial denaturation 95 °C for 5 min, followed by 35 cycles of 94 °C for 45 s, 55 °C for 60 s and 72 °C for 60 s, with final extension at 72 °C for 10 min.
Cloning and sequencing of PCR products
The PCR products were run on a 1.5% agarose gel and visualised, documented under UV Pro gel documentation unit. Bands of the expected size (~500 bp) were excised from agarose gel and were purified using a DNA gel purification kit (Sigma Aldrich). The obtained purified product was cloned into a pTZ57R/T vector (Thermo Scientific) and transformed into competent Escherichia coli DH5α cells by heat-shock method (Sambrook and Russel 2001). Total of sixty recombinant clones i.e. thirty each from the two primer combinations were randomly picked and plasmid DNA from the transformed clones was isolated using plasmid elution kit (Sigma Aldrich) and confirmed by plasmid PCR. Plasmid DNAs were sequenced for further analysis.
Sequence analysis and phylogenetic tree construction
All the isolated DNA sequences were trimmed for vector sequence contaminations and only the conserved region between P-Loop and GLPL which represents RGA was considered for further analysis. Assembly of DNA sequences and translation to the predicted amino acid sequence were performed using Transeq software (http://www.ebi.ac.uk/Tools/st/emboss_transeq/). DNAMAN software (version 8.0) was used for homology matrix between the isolated sequences and other related R-gene sequences. Identity and similarity searches of nucleotide and amino acid sequences were performed using BLAST at the National Centre for Biotechnology Information (NCBI) database, a non-redundant GenBank (http://www.ncbi.nlm.nih.gov/BLAST/). MEME software (Multiple Expectation Maximization for Motif Elicitation) (Bailey et al. 2009) was used to analyse the conserved motif regions of the predicted RGAs. The deduced amino acid sequences of the RGAs isolated from the pepper line ‘IHR 2451’, with other related plant RGAs were aligned and used for phylogenetic tree construction using MEGA (Molecular Evolutionary Genetic Analysis) version 6.0 (Tamura et al. 2013). Cluster analysis was carried out using CLUSTALX (Thompson et al. 1997) based on the neighbour joining tree (Saitou and Nei 1987) with default settings.
Results
Isolation of resistant gene analogs (RGAs)
Nucleotide binding site-encoding DNA regions were amplified using the degenerate primers in the jackpot genotype ‘IHR 2451’. Generated fragments with expected size of approximately 500 bp were purified and cloned into the pTZ57R/T vector. Thirty recombinant clones of each primer combination were randomly selected for sequencing. Cloning and characterization of these amplicons revealed that each fragment included many RGA sequences. Out of 60 clones sequenced, twenty-eight sequences which had uninterrupted ORF were considered as RGAs. Homology matrix was done for the shortlisted 28 (CaTRGA1-28) sequences from this study and five sequences (CaTRGA1, CaTRGA4, CaTRGA14, CaTRGA15 and CaTRGA18) with <61% homology were selected for further analysis and were deposited in the GenBank data base (http://www.ncbi.nlm.nih.gov/genbank) and their accession numbers are KX758993 (CaTRGA1), KX758994 (CaTRGA4), KX758995 (CaTRGA14), KX758996 (CaTRGA15) and KX758997 (CaTRGA18). The protein sequences of these RGAs were subjected to BLASTp algorithm and indicated that these RGAs had high scores compared with the RGAs or R-genes identified in other plant species (Table 2). These five sequences were compared for homology with highly characterized RGAs RPM1, Gpa2, M, N, and PRF (Table 3).
Multiple amino acid sequence alignment
The region between the P-loop and GLPL was used to achieve multiple-amino acid sequence alignment between the other plant R-genes and the deduced amino acid sequences of Capsicum RGAs. In the present study, isolated NBS-LRR was classified into TIR or non-TIR according to the characteristic amino acid at the end of the kinase-2 domain. The presence of aspartic acid (D) at the end of kinase-2 domain is classified as TIR, and according to this 13 sequences of the present study (CaTRGA1-13) belongs to TIR group along with N and M. When there is a tryptophan (W) at the end of kinase-2 domain, the sequence can be classified as non-TIR group and CaTRGA14-28 are grouped along with RPM1, Gpa2 and PRF (Fig. 1).
Multiple sequence alignment of deduced amino acid sequences of the NBS domain of 28 CaTRGAs with other plant R-genes. Major conserved motifs (P-loop, kinase 2, RNBS-B, RNBS-C and HDGLPL) are highlighted in alignment. Gaps to optimize multiple sequence alignment are indicated by (dot). The construction of multiple sequence alignment was performed by using DNAMAN software
Multiple Expectation Maximization for Motif Elicitation motif analyser was used to identify structural diversity of conserved domains and their relative position in chilli RGAs (Fig. 2I). Six highly conserved motifs (P-loop, RNBS-A, kinase-2 RNBS-B, RNBS-C and GLPL) were observed across the sequences of 28 isolated RGAs represents the NBS-LRR R-genes structural characteristics (Fig. 2II).
I Distribution of conserved motifs in the NBS domain of 28 CaTRGAs generated through MEME software tool. A different coloured box represents different conserved motifs. II Sequence signature of Six major conserved motifs in eggplant RGAs NBS region along with their E values (motif 1 kinase 2, motif 2 P-loop, motif 3 RNBS-B (kinase 3A), motif 4 RNBS-C, motif 5 HD-GLPL, motif 6 RNBS-A (Non-TIR) and motif 7 RNBS-A (TIR)
Phylogentic analysis
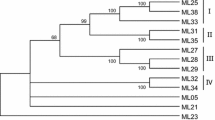
Diversity analysis of RGAs was performed with the deduced amino acid sequences of twenty-eight isolated RGAs in this study from the resistant hot pepper cultivar ‘IHR 2451’ and other RGA sequences of chilli with the known R-genes including truncated N protein (Nicotiana tabacum), root-knot nematode resistance protein M (Solanum lycoperiscum), Prf (Solanum lycoperiscum), Gpa2 (Solanum tuberosum), and RPM1 (Arabiodopsis thaliana) by use of the software MEGA 6.0 (Tamura et al. 2013) using maximum likelihood method along jones-taylor-thornton (JTT) model with 1000 bootstrap replications. The Phylogenetic tree is initially divided into two clades referring TIR and non-TIR-NBS-LRR and the results suggested wide genetic diversity of Capsicum RGAs. CaTRGA1-CaTRGA13 are grouped with highly characterized TIR-NBS-LRR R-genes called N and M. CaTRGA14-CaTRGA28 are grouped with non-TIR-NBS-LRR of PRF, Gpa2 and RPM1 (Fig. 3).
Molecular phylogenetic analysis of CaTRGAs with R-genes of different plants and R-gene representatives of TIR and non-TIR NBS-LRRs by using maximum likelihood method along with jones-taylorthornton (JTT) model. The numbers on the branches of tree are confidence values based on bootstrap method with 1000 replications
The five RGAs (CaTRGA1, CaTRGA4, CaTRGA14, CaTRGA15 and CaTRGA18) which showed high similarities with the resistant genes were compared with other sequences which include Rpi-blb2 (accession no. DQ122125), Mi-1 (AF091048), CaMi (DQ465824), SW-5F (JX026925), SW-5B (AY007366), PRF, N, M, I2 (AF118127), HRT (AF234174), tm-2 (AY765395), Tm-2^ 2, Tm-2 (AF536200), rx (AJ011801), RGC1 (AF266747), Gpa2, and Pvr-9 (KM590984 and KM590985). Neighbour joining method with MEGA6 software was used and an unrooted scaled phylogenetic tree was constructed (Fig. 4). The analysis indicated that TIR and non-TIR sequences formed different clades. Analysis showed that nematode resistant proteins (DQ465824, AF091048), Rpi-blb2, Pvr9, CaTRGA15 and CaTRGA18 formed a monophyletic clade.
Discussion
Being an important commercial and industrial spice crop, improvement of disease resistance mainly for viruses is the top priority in pepper breeding programmes. Earlier screening for virus resistance, the line ‘IHR 2451’ as jackpot genotype showed resistance to two major viruses (Naresh et al. 2016). Identification of RGAs and their use in identification of markers for disease resistance for marker assisted selection is an ideal approach (Yang et al. 2013; An and Yang 2014; Mutlu et al. 2008). These studies indicate the potentiality of R-gene analogues in identification of markers linked to disease resistance, as R-gene markers target conserved motifs of resistant genes (Joshi et al. 2012). To our knowledge, the isolated RGAs in this study are the first report of NBS-LRR class of RGAs isolated from multiple virus resistance chilli genotype and the phylogenetic results suggested wide genetic diversity of isolated Capsicum RGAs with previous studies (Fig. 3).
These NBS–LRR proteins are mainly of two types, TIR and non-TIR types both involved in pathogen recognition but are distinctive in sequence and signalling pathways (Michale et al. 2006). In our study, the cloned sequences belonged to both TIR and non-TIR–NBS–LRR type RGAs, which is similar to results reported in eggplant (Reddy et al. 2015), cucumber (Wan et al. 2010) and peppers (Wan et al. 2012) using the same degenerate primers, indicating the success of homology based PCR cloning of NBS-LRR class disease resistance gene analogs in Capsicum. Out of 28 sequences, CaTRGA1-13 sequences were TIR type having highly conserved aspartic acid (D) at the end of the kinase-2 domain and CaTRGA14-28 sequences were classified as non-TIR type as indicated by the final residue tryptophan (W) at the end of kinase-2 motif. In case of monocots only non-TIR-NBS-LRR genes are present, but in dicots the presence of both types are reported (Joshi et al. 2010). This confirms that chilli being dicotyledonous, has both TIR and non-TIR NBS-LRR class genes. In our study the TIR: non-TIR type NBS-LRR RGAs ratio of 1:1 have been found, whereas high ratios were reported in other crops (Donald et al. 2002). But in eggplant only non-TIR RGAs were isolated by Zhuang et al. (2012). Thus it indicates that the amount and ratio of TIR and non-TIR types varies across the plant species (An and Yang 2014; Reddy et al. 2015).
Out of the 28 RGAs, five RGAs which have high homology with other resistant genes were selected and these sequences were translated into protein sequences. The homology matrix results indicates that the isolated chilli RGAs have similarity with the highly characterized R-genes and were considered as NBS-LRR class of genes. These RGAs were subjected to BLASTp and indicated that these RGAs have high scores compared with the RGAs or R genes and have high identity with the plant disease resistance proteins (Table 2). Resistant gene analogs are the best way to develop PCR based markers linked to the resistant genes as these are candidates involved in plant defence mechanism (Peter et al. 2013). Major Tomato mosaic virus (ToMV) resistance have been reported to be conferred by Tm-1 gene and Kang et al. (2010) developed SNP markers for CMV resistance by comparative mapping using homolog of Tm-1 gene in tomato, as Cmr1 gene (single dominant gene conferring resistant to CMV in Bukang) is located on LG2 which is synthetic to ToMV resistance locus (Tm-1). Similarly Yang et al. (2009) isolated RGAs from pepper variety Yv2007001, a highly resistant line for CMV using degenerate primers and PCR approach and found RGA 46 showed high homology with Tm-2 2(ToMV), suggesting its correlation with CMV resistance. The RGA sequences isolated in present study showed high identity with the different resistant protein sequences in other crops. The lower E value suggests the most reliable match (Totad et al. 2005). Based on E value, CaTRGA1 showed high identity with ToMV resistance protein N-like in chilli (XP_016574092.1 99% identity with 5e−110 E value) and CaTRGA4 showed high identity with TMV resistance protein N-like in Solanum pennellii (XP_015062483.1, 85% identity with E value 2e−90), respectively. CaTRGA18 showed 51% identity with Pvr9 (Pepper veinal mottle virus resistant gene characterized from the resistant source pepper genotype CM 334) (AJW77761, E value 1e−41) indicating that these RGAs have high correlation with virus resistance. CaTRGA18 also showed 55% identity with late blight resistance protein Rpi-blb2 (Solanum bulbocastum) (AAZ95005, E value 4e−44) as it is already reported that Pvr9 gene is Rpi-blb2 ortholog (Tran et al. 2015), they also reported that Pvr9 gene putatively encodes for possible CC-NBS-LRR protein. This suggests the presence of Pvr9 gene for poty virus resistance in the ‘IHR2451’ genotype. Recently Kim et al. (2016) reported that Pvr4 (a poty virus resistant gene in C. annuum) and Tsw (Tomato spotted wilt virus resistant gene in C. chinense) encode for nucleotide binding and leucine rich repeat domain proteins and these multiple virus resistant genes diverged from a progenitor gene of a common ancestor in Capsicum spp. as indicated by genomic structure and coding sequences of these two genes. Similarly, CaTRGA15 showed high identity with root-knot nematode resistance protein in Capsicum annuum L. (ABE68835.1, 79% identity with E value 2e−83); similarly Zhang et al. (2008) isolated RGA-p20 sequence which had high similarity with Mi 1.2 gene conferring resistance to root knot nematode resistance.
Neighbour joining tree phylogentic analysis of these 5 RGAs (CaTRGA1, CaTRGA4, CaTRGA14, CaTRGA15 and CaTRGA18) with other characterized plant resistant proteins showed that nematode resistant proteins (DQ465824, AF091048), Rpi-blb2, Pvr9 (Pepper veinal mottle virus resistant gene), CaTRGA15 and CaTRGA18 formed a monophyletic clade (Tran et al. 2015). These CaTRGA15 and CaTRGA18 could be potential candidates for studying the nematode and virus resistance in chilli.
Usually in plants R-genes are present in clusters of tightly linked genes (Hulbert et al. 2001). It is already reported that the L gene occur in a cluster of resistance genes located on chromosome 11 in a position that apparently overlaps with resistance gene clusters in tomato and pepper (Grube et al. 2000b). Lefebvre (2004) reported the existence of R-gene cluster where disease resistance QTLs for potyvirus, cucumber mosaic virus (CMV), Tomato spotted wilt virus (TSWV), and Phytophthora capsici. Further, mapping of these RGAs isolated from jackpot genotype ‘IHR 2451’ is required to identify the R-gene cluster.
Conclusion
The present study identified five candidates of RGAs from a multiple virus resistant pepper genotype ‘IHR 2451’ can be used for characterization of different resistant genes. These RGAs identified could be regarded as candidate sequences of disease resistance, which may be explored in developing molecular markers for disease resistance. Mapping of these RGAs will help in better understanding of multiple virus resistance and identifying and cloning of R-genes in peppers for marker assisted breeding programs for virus resistance.
References
An H, Yang K (2014) Resistance gene analogs in walnut (Juglans regia) conferring resistance to Colletotrichum gloeosporioides. Euphytica 197(2):175–190
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:202–208
Basak J, Kundagrami S, Ghose TK, Pal A (2004) Development of yellow mosaic virus (YMV) resistance-linked DNA marker in Vigna mungo from populations segregating for YMV reaction. Mol Breed 14:375–383
Chen XM, Hayes PM, Toojinda T, Vivar H, Kudrna D, Kleinhofs A, Leung H, Line RF (1998) Genetic mapping of genes for stripe rust resistance in barley using resistance gene analog polymorphism and AFLP markers. Phytopathology 88:5–16
Deng Z, Huang S, Ling P, Chen C, Yu C, Weber CA, Moore GA, Gmiter FG (2000) Cloning and characterization of NBS-LRR class resistance gene candidate sequences in citrus. Theor Appl Genet 101:814–822
Donald TM, Pellerone F, Adam-Blondon AF, Bouquet A, Thomas MR, Dry IB (2002) Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor Appl Genet 104:610–618
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Ellis J, Dodds P, Pryor T (2000) Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol 3:278–284
Grube RC, Blauth JR, Andredos MSA, Caranta C, Jahn MK (2000a) Identification and comparative mapping of a dominant potyvirus resistance gene cluster in Capsicum. Theor Appl Genet 101:852–859
Grube RC, Zhang Y, Murphy JF, Loaiza-Figueroa F, Lackney VK, Provvidenti R, Jahn M (2000b) New source of resistance to Cucumber mosaic virus in Capsicum frutescens. Plant Dis 84:885–891
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Biol 48(1):575–607
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Johal GS, Briggs SP (1992) Reductase activity encoded by the Hm1 disease resistance gene in maize. Science 258:985–987
Joshi RK, Monhanty S, Subudhi E, Nayak S (2010) Isolation and characterization of NBS–LRR resistance gene candidates in turmeric (Curcuma longa cv surama). Genet Mol Res 9:1796–1806
Joshi RK, Mohanty S, Kar B, Nayak S (2012) Assessment of genetic diversity in Zingiberaceae through nucleotide binding site-based motif-directed profiling. Biochem Genet 50:642–656
Kanazin V, Marek LF, Shoemarker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Kang BC, Yeam I, Jahn MM (2005) Genetics of plant virus resistance. Annu Rev Phytopathol 43:581–621
Kang WH, Hoang NH, Yang HB, Jin-Kyung Kwon, Jo Sung-Hwan, Jang-Kyun Seo, Kim KH, Choi D, Kang BC (2010) Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L). Theor Appl Genet 120:1587–1596
Kim SB, Kang WH, Huy HN, Yeom SI, An JT, Kim S, Kang MY, Kim HJ, Jo YD, Ha Y, Choi D (2016) Divergent evolution of multiple virus-resistance genes from a progenitor in Capsicum spp. New Phytol. doi:10.1111/nph14177
Lefebvre V (2004) Molecular markers for genetics and breeding: development and use in pepper (Capsicum annuum L). In: Lorz H, Wenze G (eds) Biotechnology in agriculture and forestry molecular marker systems. Springer, Heidelbeg, pp 189–214
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P (1998) Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA 95:370–375
Michale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS–LRR proteins: adaptable guards. Genome Biol 7:212
Mutlu N, Boyac FH, Gocmen M, Abak K (2008) Development of SRAP, SRAP-RGA, RAPD and SCAR markers linked with a Fusarium wilt resistance gene in eggplant. Theor Appl Genet 117:1303–1312
Naresh P, Krishna Reddy M, Hema Chandra Reddy P, Madhavi Reddy K (2016) Screening chilli (Capsicum spp.) germplasm against Cucumber mosaic virus and Chilli veinal mottle virus and inheritance of resistance. Eur J Plant Pathol 146(3):451–464
Noir S, Combes MC, Anthony F, Lashermes P (2001) Origin, diversity and evolution of NBS-type disease-resistance gene homologues in coffee trees (Coffea spp. L). Mol Genet Genomics 265:654–662
Pallas V, Garcia JA (2011) How do plant viruses induce disease? Interactions and interference with host components. J Gen Virol 92:2691–2705
Peter P, Ildiko V, Maarja L, Andras C, Neil B, Jari PTV, Jaakko H (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9:6
Reddy AC, Venkat S, Singh TH, Aswath C, Reddy KM, Reddy LDC (2015) Isolation, characterization and evolution of NBS-LRR encoding disease-resistance gene analogs in eggplant against bacterial wilt. Eur J Plant Pathol 143(3):417–426
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbour Laboratory Press, Cold Spring Harbour
Sharma TR, Das A, Kumar SP, Lodha ML (2009) Resistance gene analogues as a tool for rapid identification and cloning of disease resistance genes in plants: a review. J Plant Biochem Biotechnol 18(1):1–11
Singh J, Thakur MR (1979) Genetics of resistance to tobacco mosaic virus, cucumber mosaic virus and leaf curl virus in hot pepper (Capsicum annuum L). In: 3rd Eucarpia meeting, Montfavet, pp 119–123
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Totad AS, Fakrudin B, Kuruvinashetti MS (2005) Isolation and characterization of resistance gene analogs (RGAs) from sorghum (Sorghum bicolor L Moench). Euphytica 143:179–188
Tran PT, Choi H, Choi D, Kim KH (2015) Molecular characterization of Pvr9 that confers a hypersensitive response to Pepper mottle virus (a potyvirus) in Nicotiana benthamiana. Virology 481:113–123
Wan HJ, Zhao ZG, Malik AA, Qian CT, Chen JF (2010) Identification and characterization of potential NBS-encoding resistance genes and induction kinetics of a putative candidate gene associated with downy mildew resistance in Cucumis. BMC Plant Biol 10:186
Wan H, Yuan W, Ye Q, Wang R, Ruan M, Li Z, Zhou G, Yao Z, Zhao J, Liu S, Yang Y (2012) Analysis of TIR- and non-TIR-NBS-LRR disease resistance gene analogous in pepper: characterization, genetic variation, functional divergence and expression patterns. BMC Genomics 13:502
Yang XL, Wang SB, Liu JB, Pan BG, Wan HJ, Chen JF (2009) Analysis and cloning of Cucumber mosaic virus resistance gene analogs in pepper (Capsicum annuum L). Jiangsu J Agric Sci 4:26
Yang L, Li D, Li Y, Gu X, Huang S, Garcia-Mas J, Weng Y (2013) A 1,681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol 13:53
Yi Y, Li R, Xu H, Wu X, Li S, Zhang J, Yin Y (2013) Identification of SRAP and RGA markers linked to the powdery mildew (Blumeria graminis) resistance gene PmZB90 in common wheat. Aust J Crop Sci 7:454–459
Zhang L, Chen R, Zhang J, Ouyang B, Xiao J, Li H, Ye Z (2008) Cloning and analysis of resistance gene analogs from pepper (Capsicum annuum L). Scientia Agricultura Sinica 41(1):169–175
Zhuang Y, Zhou X, Wang S (2012) Genetic diversity of NBS–LRR class disease-resistance gene analogs in cultivated and wild eggplants. Plant Syst Evol 298(7):1399–1406
Acknowledgements
Authors acknowledge The Director, ICAR-Indian Institute of Horticulture Research, Bangalore for providing research facilities and Department of Science and Technology, Government of India for providing fellowship for Ph.D. studies of Dr. P. Naresh.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Naresh, P., Krishna Reddy, M., Reddy, A.C. et al. Isolation, characterization and genetic diversity of NBS-LRR class disease-resistant gene analogs in multiple virus resistant line of chilli (Capsicum annuum L.). 3 Biotech 7, 114 (2017). https://doi.org/10.1007/s13205-017-0720-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-0720-y