Abstract
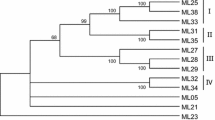
Degenerate primers designed based on known resistant genes (R-genes) and resistance gene analogs (RGAs) were used in combinations to elucidate RGAs from Sorghum bicolor, cultivar M 35-1. Most of the previously tried primer combinations resulted in amplicons of expected 500–600 bp sizes in sorghum along with few novel combinations. Restriction analysis of PCR amplicons of expected size revealed a group of fragments present in a single band indicating the heterogeneous nature of the amplicon. Many of these were cloned and some were considered for analysis. The nucleotide sequence of different cloned fragments was done and their predicted amino acid sequences compared to each other and to the amino acid sequences of known R-genes revealed significant sequence similarity. A cluster analysis based on neighbor-joining (N-J) method was carried out using sorghum RGAs (SRGAs) together with several analogous known R-genes resulting in two major groups; cluster-I comprising only SRGAs and cluster-II comprised of known R-gene sequences along with three SRGAs. Further analysis clearly indicated similarity of SRGAs in overall sense with already known ones from other crop plants. These sequences can be used as guidelines to detect, map and eventually isolate numerous R-genes in sorghum.
Similar content being viewed by others
References
Aarts, M.G.M., B.L. Hekkert, E.B. Holub, J.L. Beynon, W.J. Stiekema & A. Pereira, 1998. Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in A. thaliana. Mol Plant-Microbe Interact 11: 251–258.
Altschul, S.F., T.L. Medden, A.A. Schaffer, J. Zhang, W. Miller & D.J. Lipman, 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programmes. Nucl Acids Res 25: 3389–3402.
Altschul, S.F., W. Gish, W. Miller, E.W. Myers & D.J. Lipman, 1990. Basic local alignment search tool. J Mol Biol 215: 403–410.
Baker, B., P. Zambryski, B. Staskawicz & S.P. Dinesh-Kumar, 1997. Signaling in plant-microbe interactions. Science 276: 726–733.
Collins, N.C., C.A. Webb, S. Seah, J.G. Ellis, S.H. Hulbert & A. Pryor, 1998. The isolation and mapping of disease resistance gene analogs in maize. Mol Plant-Microbe Interact 11: 968–978.
Dixon, M.S., D.A. Jones, J.S. Keddie, C.M. Thomas, K. Harrison & J.D.G. Jones, 1996. The tomato cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84: 451–459.
Dixon, M.S., K. Hatzixanthis, D.A. Jones, K. Harrison & J.D.G. Jones, 1998. The tomato cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine rich repeat copy number. Plant Cell 10: 1915–1925.
Falsenstein, J., 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791.
Flor, H.H., 1956. The complementary genic system in flax and flax rust. Adv Genet 8: 29–54.
Gassmann, W., M.E. Hinsh & B.J. Staskawicz, 1999. The Arabidopsis RPS4 bacterial resistance gene is a member of the TIR-NBS-LRR family of disease resistance gene. Plant J 20: 265–277.
Gebhardt, C., 1997. Plant genes for pathogen resistance-variation on a theme. Trends Plant Sci 2: 243–244.
Grant, M., L. Godiard, E. Straube, T. Ashfield, J. Lewald, A. Sattler, R. Innes & J. Dangl, 1995. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846.
Hall, T.A., 1999. BioEdit: A user-friendly biological sequences alignment editor and analysis program for Windows 95/98/NT. Nucl Acid Symp Ser 41: 95–98.
Hammond-Kosack, K. & J.D.G. Jones, 1997. Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48: 575–607.
Johal, G.S. & S.P. Briggs, 1992. Reductase activity encoded by the Hm1 disease resistance gene in maize. Science 258: 985–987.
Jones, D.A., C.M. Thomas, K.E. Hammond-Kosack, P.J. Balint-Kurti & J.D.G. Jones, 1994. Isolation of the tomato cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793.
Kanazin, V., F.L. Marek & R.C. Shoemaker, 1996. Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93: 11746–11750.
Lawrence, G.J., E.J. Finnegan, M. Ayliffe & J. Ellis, 1995. The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7: 1195–1206.
Leister, D., A. Ballvora, F. Salamani & C. Gebhardt, 1996. A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14: 421–429.
Lopez, C.E., I.F. Acosta, C. Jara, F. Pedraza, E. Gaitan-Solis, G. Gallego, S. Beebe & J. Tohme, 2002. Identifying resistance gene analogs associated with resistances to different pathogens in common bean. Phytopathology 93: 88–95.
Mago, R., S. Nair & M. Mohan, 1999. Resistance gene analogs from rice: Cloning, sequencing and mapping. Theor Appl Genet 99: 50–57.
Madsen, L.H., N.C. Collins, M. Rakwalska, G. Backes, N. Sandal, L. Krusell, J. Jensen, E.H. Waterman, A. Jahoor, M. Ayliffe, A.J. Pryor, P. Laugridge, P. Schulze-Lefert & J. Stougaard, 2003. Barley disease resistance gene analogs of the NBS-LRR class: Identification and mapping. Mol Gen Genomics 269: 150–161.
Martin, G.B., S.H. Brommonschenkel, J. Chunwongse, A. Grary, M.W. Ganal, R. Spivey, T. Wu, E.D. Earle & S.D. Tanksley, 1993. Map based cloning of protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436.
Maunder, A.B., 1993. Breeding for stalk rot resistance as a component of acceptable agronomic performance. In: Proceedings of the Consulative Group Discussion of Research Needs and Strategies for Control of Sorghum Root and Stalk Rot Diseases Sorghum Root and Stalk Rots, Critical Review, 27 November–2 December 1983, Bellagio, Italy. ICRISAT, Patancheru, Andhra Pradesh 502324, India, pp. 219–224.
Murray, M.G. & W.F. Thomson, 1980. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8: 4321–4325.
Pan, Q., J. Wendel & R. Fluhr, 2000. Divergent evolution of plant NBS-LRR resistance gene homologs in dicot and cereal genomes. J Mol Evol 50: 203–213.
Parker, J.E., M.J. Coleman, V. Szabò, L.N. Frost, R. Schmidt, E.A. van der Biezen, V. Moore, C. Dean, M.J. Daniels & J.D.G. Jones, 1997. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and Inerleukin-1 receptors with N and L6. Plant Cell 9: 879–894.
Quint, M., C.M. Dussle, A.E. Mechinger & T. Luebberstedt, 2003. Identification of genetically linked RGAs by BAC screening in maize and implications for gene cloning, mapping and MAS. Theor Appl Genet 106: 1171–1177.
Saitou, N. & M. Nei, 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425.
Salmeron, J.M., G.E.D. Oldroyd, C.M.T. Rommens, S.R. Scofield, H.S. Kim, D.T. Lavelle, D. Dahlbeck & B.J. Staskawicz, 1996. Tomato Prf is a member of leucine-rich repeats class of plant disease resistance gene and lies embedded within the Pto kinase gene cluster. Cell 86: 123–133.
Shen, K.A., V. Meyers, M.N. Islani-Faridi, D. Chin, B. Stelly & R.W. Michelmore, 1998. Resistance gene candidates identified by PCR with degenerate oligonucleotide primer map to clusters of resistance genes in lettuce. Mol Plant-Microbe Interact 8: 815–823.
Song, W.Y., G.L. Wang, L.L. Chen, H.S. Kim, L.Y. Pi, T. Holsten, J. Gardner, B. Wang, W.X. Zhai, L.H. Zhu, C. Fanquet & P.C. Ronald, 1995. A receptor kinase like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806.
Soriano, J.M., S. Vilanova, C. Romero, G. Llácer & M.L. Badenes, 2005. Characterization and mapping of NBS-LRR resistance gene analogs in apricot (Prunus armeniaca L.). http://www.springerlink.com/media/B5UC1606PK4YWLAC5J 7W/Contributions/8/7/9/C/879C924T9H6K1KH3_html/fulltext. html.
Thomas, C.M., D.A. Jones, M. Parniske, K. Harrison, P.J. Balint-Kurti, K. Hatzixanthis & J.D.G. Jones, 1997. Characterization of the tomato cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognition specificity in cf-4 and cf-9. Plant Cell 9: 2209–2224.
Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin & D.G. Higgins, 1997. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acid Res 25: 4876–4882.
Whitham, S., S.P. Dinesh-Kumar, D. Choi, R. Hehl, C. Corr & B. Baker, 1994. The product of the tobacco mosaic virus resistance genes N: Similarity to Toll and the interleukin-1 receptor. Cell 78: 1101–1115.
Yu, Y., G.R. Buss & M.A. Maroof, 1996. Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93: 11751–11756.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Totad, A.S., Fakrudin, B. & Kuruvinashetti, M.S. Isolation and characterization of resistance gene analogs (RGAs) from sorghum (Sorghum bicolor L. Moench). Euphytica 143, 179–188 (2005). https://doi.org/10.1007/s10681-005-3428-8
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s10681-005-3428-8