Abstract
Known as the Roscoff worm or mint-sauce worm, Symsagittifera roscoffensis is an Acoel distinguishable due to the presence of symbiotic alga Tetraselmis convolutae, held beneath the epidermis. Isolated populations of S. roscoffensis span a broad geographical range along the north-eastern Atlantic coast, from Wales to Portugal. The only known population of the worm in the United Kingdom was discovered in Wales decades ago, but very little research has been conducted since. For 13 months, we measured how environmental conditions such as temperature, salinity and light intensity coincided with population size at the Welsh field site. To establish phylogenetic relationships among the different populations and their algal symbionts, we designed new polymerase chain reaction (PCR) oligonucleotides to assess the nucleotide diversity of the mitochondrial cytochrome c oxidase I subunit (COI) gene in gDNA extracted from representative worms across their known range (Wales, France, Portugal, Spain, and Guernsey). We also targeted the 18S rRNA gene of their algal symbiont, Tetraselmis convolutae. We observed temporal shifts in environmental factors coinciding with fluctuating worm colony size, notably temperature. Based on the molecular data, the worm exhibited different ecotypes across locations, while the algal symbiont showed little genetic variation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Symsagittifera roscoffensis (previously Convoluta roscoffensis) is an Acoel in the phylum Xenacoelomorpha (previously Platyhelminthes; ITIS 2019), and lives within the intertidal zone. First taxonomically described over a century ago, it was termed a “plant animal” due to its symbiotic relationship with the chlorophyte alga Tetraselmis convolutae (von Graff 1891). An adult is 2–4 mm in length, lacks a defined coelom (Bailly et al. 2014), and is characterised by a vivid green colour due to its algal symbiont (Douglas 1983; Arboleda et al. 2018). The relationship between T. convolutae and S. roscoffensis provides the host with all of its nutritional needs and no heterotrophic feeding is known to take place (Bailly et al. 2014; Arboleda et al. 2018; Thomas et al. 2023a). While symbiosis can occur with other members of the Genus Tetraselmis in the laboratory, suboptimal algal species can result in increased mortality (Arboleda et al. 2018; Thomas et al. 2023b). Aposymbiotic juveniles acquire algal cells from the environment, as they do not transfer vertically via the parental line (Bailly et al. 2014; Provasoli et al. 1968).
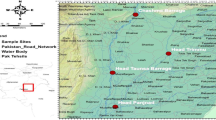
Since it was first discovered in Roscoff in 1879 (Geddes 1879), populations of S. roscoffensis have been reported in Wales, France, the Channel Islands, Spain and Portugal (Jondelius et al. 2011; Carvalho et al. 2013; Bailly et al. 2014; Franks et al. 2016; Mettam 1979). The intertidal zone, where S. roscoffensis resides, is a dynamic environment. Inhabitants are often exposed to prevailing weather conditions during low tide, and surrounding conditions can change rapidly as a result. The only sighting of the worm in the UK was in Limpet Bay in East Aberthaw, Wales (Mettam 1979). The beach is rocky with limited open patches of sand; the rocks are primarily limestone arising from the falling cliff-face of the Jurassic coastline (Fig. 1A and B). Open patches of sand are rare on the beach, instead clear spaces are dominated by thick clay and no sand (in some places the rocks are upon the clay with no visible sand underneath). Rocks in the area where worms are found are also small and have been eroded by the sea. Larger rocks are in place behind S. roscoffensis’ habitat closer to the cliff face. Symsagittifera roscoffensis is found in the small pools of water, between these rocks where the underlaying substrate is sand. Upon discovery of a population at this Welsh site, Mettam (1979) speculated that S. roscoffensis had made its way up the Bristol Channel from the nearest population in the Channel Islands; however, there has been no genetic data on the intra-specific diversity of S. roscoffensis to confirm or refute this hypothesis.
Locations of Symsagittifera roscoffensis populations used in the present study. Symsagittifera roscoffensis populations exist along the Atlantic cost (A). Limpet Bay (red), East Aberthaw (Wales, UK) represents our study site; green, Alderney, Guernsey; blue, Costa da Caparica, Portugal; yellow, Roscoff, France. (B) Image of Limpet Bay (Wales, UK). Worms are located along the tide mark where there is a distinctive colour change in the rocks (from intertidal to supratidal)
Newly emerged worms are aposymbiotic, i.e. lacking the T. convolutae symbiont. Survival and successful establishment of a new colony requires the worms to acquire the algal symbionts post-hatching (Oschman 1966; Douglas and Gooday 1982). Symbiotic T. convolutae may travel with the founder worm population and be released upon the worm’s death and establish a local supply of algal cells or may already be present in the substrate upon the worm’s arrival. The ability of the worms to acquire the algae externally from the environment may lead to distinct clades of T. convolutae being associated with different populations of S. roscoffensis (Riewluang and Wakeman 2023), it may also be the case that alternative symbionts are associated with distinct populations. The first to suggest this was Mettam (1979) and Mcfarlane (1982), stating that individuals of the Welsh site indeed formed a symbiotic relationship with a different local species of Tetraselmis, with early results suggesting that the alternative symbiont could be found in up to 55% of individuals. The variation between the preferred symbiont T. convolutae and an alternative symbiont reported by Mettam (1979) and Mcfarlane (1982) was dependent upon its location along the intertidal zone. However, there were no reports of mixed symbionts within a single worm. The authors distinguished the different algal symbionts by microscopy, based on differences in the shape of the pyrenoid between species. Macfarlane (1982) suggested the reason for differences in symbiont profiles was due to the preferred Tetraselmis being less abundant at the Welsh field site.
The site in Wales is the most northerly location of S. roscoffensis’ known distribution (Mcfarlane 1982), but the literature provides scant information regarding this colony. To address this knowledge gap, we measured environmental parameters and population characteristics of the worm at the Welsh site for a period of 13 months. Secondly, we isolated worm DNA from geographically distinct populations (Wales, Guernsey, France, Portugal) and used the cytochrome c oxidase I gene to determine population relatedness. Lastly, using the same worm extracts, we probed DNA from the algal symbionts of S. roscoffensis collected from Wales, Guernsey, France and Portugal using the 18S rRNA gene to confirm the identity of the algal symbiont.
2 Methods
2.1 Field sampling
Data collection started in August 2020 and ended in August 2021. Twice per month we collected measurements at the field location in East Aberthaw, Wales (GIS:51.38158, -3.36363). We selected six colonies along a transect at the habitat range of S. roscoffensis; each colony was assigned a permanent marker point. We then returned to the same point each time to measure colony size, temperature, salinity, and light intensity. During the entire study period (13 months), and subsequent visits for sample collections and observations, we noted that low or high tides did not seem to cover the colonies at any point.
About 50 ml of sea water was collected as close as possible to each marker, and the water temperature was measured immediately with a thermometer (Silverline digital). Salinity was measured with a refractometer (D-D True Seawater). Light intensity was measured using an Apogee MQ-500 Quantum meter placed directly above the worm colony. Environmental data are presented as mean ± SE for each month (at least 6 technical measures were taken across the site on a given sampling day).
Pictures of each colony were taken alongside a reference object for scale, images were processed using Image J software. We set the scale in Image J based on the reference object in the picture, then measured the size of each colony as surface area. The individual colony size measurements were summed to calculate the cumulative colony size, yielding two cumulative colony size measurements per month. Population size data are expressed as cumulative colony size ± SE for each month.
2.2 Worm collection and DNA extraction
Samples of S. roscoffensis collected from France, Guernsey, and Portugal (Fig. 1A) were preserved in 70% ethanol and stored at -80 °C upon arrival and prior to gDNA extractions. Samples from Wales were collected at East Aberthaw (see Sect. 2.1), maintained temporarily using culture conditions described by Thomas et al. (2023a), salinity of 30 enriched with 10 ml per L of Guillards solution (Guillard and Ryther 1962) (F/4), 14.5 ºC, 16 L/8D, and 69 µmol m− 2 s− 1. DNA was extracted from live worms collected from the Welsh field site only.
Worms suspended (approx. 50 individuals) in 1.5 ml of F/4 medium, seawater or ethanol were centrifuged at 10 x g for 10 min at room temperature (~ 22 °C); the pellet was retained, and the supernatant was discarded. Each preparation was probe for both the worm (cox1) and algal (18S rRNA) gene targets. Genomic DNA was extracted using a Qiagen blood and tissue kit (https://www.qiagen.com). The manufacturer’s protocol was followed with a minor amendment to the lysis time: Fresh samples were incubated at 56 °C for 10 min with 15s vortexing every 5 min. For each of the samples preserved in 70% EtOH and frozen at -80 °C, the incubation time was increased to 1 h with intermittent vortexing for a maximum duration of 15s. Post gDNA purification, elutants were assessed for potential contaminants (salt, protein) using the Nanodrop Spectrophotometer.
2.3 Targeting the cytochrome c oxidase I and 18S rRNA genes
Amplification of both genes was achieved using end-point PCR. For the host S. roscoffensis, we targeted the cytochrome oxidase I gene (cox1), whereas for the algal symbiont, we targeted the 18S ribosomal RNA gene (18S rRNA). Cox 1 amplification was performed using newly designed oligonucleotide primers (synthesised by Eurofins, Ebersberg, Germany): Forward, 5′-GCTTATAATGTGGTRATTACTGCTC–3′, and Reverse, 5′-CAGTAAGAAGTATTGTAATACCTCCTGC-3′. These primers were selected following multiple alignment (Clustal Omega; https://www.ebi.ac.uk/Tools/msa/clustalo/), and scrutiny, of available sequences for S. roscoffensis (HM233750, FR837904) and Convolutriloba retrogemma (EU710942, EU710925) in GenBank. Each PCR reaction was carried out in a total of 25 µL using 2X Master Mix (New England Biolabs), containing 1.5 mM MgCl2, 0.2 mM dNTPs, 25 units/ml Taq DNA Polymerase, 1 µl of each primer at 10 mM working stock and ~ 190 ng template DNA per reaction. Thermocycling conditions consisted of an initial denaturation step of 94 °C for 2 min, followed by 34 cycles of 94 °C for 30 s, 57 °C for 1 min and 72 °C for 1 min, prior to a final extension step of 72 °C for 5 min (post PCR, samples were stored at 4 °C).
The algal 18S rRNA gene was amplified using published primers and thermocycling conditions; Forward: 5′-GCGGTAATTCCAGCTCCAATAGC–3′ and Reverse: 5′-GACCATACTCCCCCCGGAACC-3′ (Lim et al. 2012). PCR reactions were carried out in a total of 25 µL reaction volume using 2X Master Mix (New England Biolabs), containing 1.5 mM MgCl2, 0.2 mM dNTPs, 25 units/ml Taq DNA Polymerase, 1 µl of each primer at 10 mM working stock and ~ 152 ng template DNA per reaction. An initial denaturation step of 94 °C for 5 min, followed by 30 cycles of 95 °C for 30 s, 57 °C for 30 s and 72 °C for 1 min, prior to a final extension step of 72 °C for 10 min (post PCR, samples were stored at 4 °C).
PCR-derived amplicons of the expected size for cox1 (478 bp) and 18S (549 bp) were confirmed using 2% (w/v) agarose gel electrophoresis and a NZYDNA ladder V (www.nzytech.com) ranging from 100 to 1000 bp. Both the cox1 and 18S rRNA gene targets were cleaned-up prior to sequencing using a Thermo-Scientific EXOSAP kit, post amplification DNA yields were confirmed using a Qubit fluorometer (Thermo Fisher Scientific). Samples were sent to Eurofins genomics (Ebersberg, Germany) for Sanger sequencing using both forward and reverse reactions.
2.4 Sequence identity and phylogenetic analyses
Eight cox1 sequences (two per worm) were used to construct consensus sequences. For the algae, 14 samples were sent for sequencing (18S rRNA), but two failed to produce reliable data. All resolved DNA sequences were inspected and trimmed manually of their primer regions. BLASTn for the 18S rRNA gene and BLASTX for the cox1 gene search algorithms (Altschul et al. 1990) were used to confirm sequence identities (top three matches for each are listed in Tables 1 and 2). For the cox1 target, we gathered a broader set of 24 reference sequences from the Convolutidae (GenBank), which were added to our newly generated sequences herein (GenBank: OQ536360 to OQ536363), yielding 28 in total, each spanning ~ 424 nucleotides (or 140 amino acids) (See Table 3). For the 18S rRNA target, 51 reference sequences retrieved from GenBank covering multiple algal genera (e.g., Tetraselmis, Dunaliella, Chlorella) were added to our new sequences (GenBank: OQ538146 to OQ538151), yielding 57 in total (See Table 3).
Multiple sequence alignments for the cox1 and 18S rRNA datasets were performed in MEGA11 using the MUSCLE function (Tamura et al. 2021). Evolutionary reconstructions were performed using the maximum likelihood method (1,000 bootstrap re-samplings) based on either the Tamura 3-perameter model (Tamura 1992) for cox1 or the Kimura 2-parameter model for 18S rRNA (Kimura 1980). DNA substitution models were selected based on the ranked Bayesian information criteria via ModelFinder in MEGA11.
Additionally, the same multiple sequence alignments for cox1 and 18S rRNA were used to reconstruct Bayesian trees in BEAST (v2.6.7); based on a yule model and MCMC chain length of 10,000,000. We used a burn in rate of 10% to summarise the posterior sample of our trees to produce the maximum clade credibility tree in tree annotator v2.6.7. FigTree software (v 1.4.4 http://tree.bio.ed.ac.uk/software/figtree/) was used to visualise the initial outputs. Final trees were formatted in iTOL (Letunic and Bork 2019) using the Bayesian generated topologies (outputs from both maximum likelihood and Bayesian inference were in good agreement).
2.5 Statistical analysis
For the environmental data (colony size, temperature, light intensity, and salinity), statistical analysis was performed using a binomial generalised linear model. Normality was confirmed using the R package DHARMa version 4.1.3 (R studio version 1.41717) that uses the Kolmogorov-Smirnov test.
3 Results
3.1 Population size and environmental conditions
The population of Symsagittifera roscoffensis at the Welsh site was at its largest during the spring months, peaking in May with 489.7 ± 27.8 cm2 (accumulative ± SE; Fig. 2). During May, water temperature was 19.7 ± 0.2 °C (mean ± SE) while salinity was 22 ± 0.7 and light intensity was 1007.4 ± 228.7 µmol m− 2 s− 1. The smallest colony sizes were observed during summer, 28.05 ± 3.1 cm2 in July, when water temperature was at its highest (27.2 ± 0.8 °C), while salinity was 24 ± 1.6 and light intensity was 1338 ± 169 µmol m− 2 s− 1. Throughout the winter, colony sizes decreased gradually: 206.15 ± 21.6 cm2 in December, 140.05 ± 22.4 cm2 in January and 125 ± 23.6 cm2 in February. Temperature in the winter months ranged from 13.3 to 5.3 °C, salinity was between 23 and 14, while light intensity was between 96.19 and 26.54 µmol m− 2 s− 1. Using binomial generalized linear models (GLMs), the variation in colony sizes between months was deemed significant, and temperature was ranked as a significant predictor variable associated with the colony size of S. roscoffensis (Table 4).
Data from the field site in East Aberthaw, Wales for (A) cumulative colony size (accumulative ± SE) of Symsagittifera roscoffensis, (B) salinity (mean ± SE), (C) water temperature (mean ± SE) and (D) ambient light intensity (mean ± SE), from August 2020-August 2021. There were significant differences between months and concerning the effect of temperature on colony sizes (Binomial, Generalized linear model; P < 0.05). Salinity and light intensity did not have a significant effect on colony sizes (GLM; P > 0.05)
3.2 Phylogenetic analyses of worm ecotypes and their algal symbionts
3.2.1 S. roscoffensis
BLASTX searches of the cox1 sequence amplified from the Welsh worms shared ~ 90% identity to S. roscoffensis collected from Spain in 2010 (GenBank acc. No. FR837904.1). Worms from Portugal, Guernsey and those re-sequenced from France were ~ 88 to > 90% similar to the same sequence from Spain (FR837904.1). In a recent publication of the genome of S. roscoffensis, Martinez et al. (2023) stated that the population of the worms have a high level of heterozygosity; however, they did not reassess the mitochondrial genome. Independent evolutionary analyses of the cox1 gene from the Convolutidae – using both maximum likelihood (ML) and Bayesian approaches – yielded trees of near identical topology (Fig. 3). All worm sequences we generated from this study, and existing sequences for France (Roscoff) and Spain (Galiza), formed a highly supported clade with 99% ML bootstrap support and a Bayesian posterior probability (BPP) of 1 (Fig. 3). This clade bifurcated (BBP = 0.99) between the sequence from Portugal (OQ536361) and all the other sampling locations (Fig. 3), and further separated the sequence from Spain (FR837904; BBP = 0.99) to those from Wales, France, and Guernsey.
Bayesian phylogenetic tree of the partial cytochrome c oxidase subunit 1 gene from marine Acoel worms (Convolutidae). Bayesian posterior probability (BPP) > 0.5 and maximum likelihood (ML) bootstrap support > 70 (from 1000 resamplings) are placed beside the respective node. The tree is rooted with a COI sequence from Daku woorimensis (FR837854). In total 28 nucleotide sequences were used for reconstructions, with four of those generated from the present study (GenBank: OQ536360 to OQ536363). Coloured circles indicate sample locations for Symsagittifera roscoffensis. The scale bar represents nucleotide substitutions per site
Notably, the two sequences from Roscoff were not identical, one is 10 years old (NC014578), and the other we generated for this study (OQ536363). Closer inspection of the nucleotide sequences revealed three (A to G) substitutions, i.e., transitions (Supp. Figure 1A), which coincided with two hydrophobic amino acid substitutions (i.e., methionine to isoleucine), and one lysine to serine substitution (Supp. Figure 1B). Looking at all the cox1 data, transitions were the most frequent single point mutations, as expected (Supp. Figure 2).
3.2.2 Algal symbionts (Tetraselmis spp.)
The 18S rRNA sequences retrieved from algal symbionts in the Welsh worms had 100% similarity to Tetraselmis sp. (MK542679.1) from Roscoff, France (2019 sample). Resident algae from worms located in Portugal, Roscoff and Guernsey shared 99.6–100% sequence identity to T. convolutae (KT860914.1). Algae initially extracted from living S. roscoffensis and subsequently grown in culture at Swansea University for ~ 6 months had a similarly high identity (99.8%) to T. convolutae (KT860914.1) – again, from Roscoff, France (2015). Interestingly, the partial 18S rRNA gene from T. convolutae – an archived sample from CCAP66/36 – shared 100% similarity to Tetraselmis sp. SMS19 (MT489380.1).
Independent evolutionary analyses of 18S rRNAs gathered from diverse algal genera, using both maximum likelihood (ML) and Bayesian approaches, produced trees with consistent topology (Fig. 4). The genus Tetraselmis was highly supported, distinct to Dunaliella (ML = 81%; BPP = 1), and both of which formed an independent clade to Chlorella (ML = 99%; BPP = 1). The sequences isolated from France, Portugal, Guernsey, and Wales formed a large, highly supported clade (BPP = 0.99) with T. convolutae, some uncultured species, and one T. astigmatica sequence (JN376804.1). These data clearly indicated the algal symbionts of worms from all locations represent Tetraselmis convolutae. Interestingly, both 18S rRNA sequences from the culture collection (T. convolutae CCAP66/36) and Swansea University short-term culture formed a diverse clade (BPP = 0.99) with all the other Tetraselmis species; T. striata, T. gracilis, T. chuii and T. suecica (Fig. 4), indicating some potential contamination from sub-culturing, or the presence of an algal consortium associated with S. roscoffensis.
Bayesian phylogenetic tree of the partial 18S rRNA gene from algae. Bayesian posterior probability (BPP) > 0.80 and maximum likelihood (ML) bootstrap support > 70 (from 1000 resamplings) are placed beside the respective node. The tree is rooted with the 18S rRNA sequence from Pavlova lutheri AC538 (JF714238). In total 57 nucleotide sequences were used for reconstructions, with six of those generated from the present study (GenBank: OQ538146 to OQ538151). Coloured circles indicate sample locations for Symsagittifera roscoffensis (containing algal symbionts). The scale bar represents nucleotide substitutions per site. The black lines, continuous and broken, represent two putative subclades within the Tetraselmis genus
4 Discussion
We completed a 13-month field campaign monitoring the environmental conditions of S. roscoffensis colonies in the least studied population of its known distribution, i.e., Wales, UK. Additionally, we gathered DNA from Welsh worms as well as those from other populations and assessed the genetic relatedness between populations (using cox1) and its algal symbiont (using 18S rRNA).
4.1 Environmental influences on S. roscoffensis colonies
Originally we expected to see the largest colony size during the summer months (Arboleda et al. 2018; Bailly et al. 2014; Douglas 1985), as this would be the period of maximum photosynthetic activity, hence growth of the worms. We also expected that during the winter months, there would be few to no worms because of the harsh environmental conditions. Our findings showed that during the winter months when both water temperature and light intensity dropped precipitously (below 10 °C and below 100 µmol m− 2 s− 1, respectively), colony sizes decreased but remained at sizeable numbers at the Welsh site (Fig. 2), suggesting that the worms were able to survive the winter condition and remain active.
Contrary to our expectation, during the summer months, population sizes reduced to lower levels than those observed in the winter months. During this period, the worms were exposed to ambient monthly average water temperature reaching > 27°C, for comparison the average ocean temperature for the same month was 17.7°C (Accessed: 26-10-2023. World Sea Temperatures 2023) The worms also experienced very high light intensities (> 1000 µmol m− 2 s− 1) that could have resulted in photoinhibition (Androuin et al. 2020). At low tides, the worms were often trapped in small pools of water only 2–10 cm deep and 1–40 cm wide; these small water bodies offered little buffering capacity against environmental stresses. Multiple worm colonies were present along the edge of the supratidal zone at the Welsh Site (Fig. 1B). During field sampling, we noted that the average incoming tide at this location did not reach the supratidal zone; therefore, these colonies would be exposed to extended periods of temperature or osmotic stresses and photoinhibition during photosynthetically active hours. While colonies of the worm would not normally survive without additional water from the incoming tide (i.e., desiccation), the unique location of the Welsh population means that it is situated in front of a saltmarsh. This could act as a potential saltwater source keeping the worms’ substrate wet in the absence of tidal water. We speculate that this could be the reason why the Welsh population is limited to one area of the beach.
Our field data indicated that colony sizes increased steadily during the spring months (March-May; Fig. 2). During this period, salinity varied little and temperature was between 15 and 20 °C, which was comparable to the reported optimal temperature for the worm in laboratory settings (10–20 °C; Thomas et al. 2023a). Salinity did not seem to have an overall effect on colony sizes in situ, which complements our previous work on worm photosynthetic output in vitro (salinity variation from 20 to 40 had little effect on photosynthetic rates; Thomas et al. 2023a). It is unsurprising that organisms living within the intertidal zone are adapted to deal with fluctuating salinity. Light intensity steadily increased from < 100 to ~ 1000 µmol m− 2 s− 1, which would have allowed the worm to increase photosynthetic activity and growth, thereby increasing their abundance. However, once temperatures exceeded 20 °C, light intensity exceeded 1200 µmol m− 2 s− 1 in the summer months (June-August), colony sizes decreased sharply (Fig. 2), suggesting that the environmental conditions became too stressful for the worm.
From our BLR model, temperature was ranked as the most likely predictor of colony size. A decrease in temperature during the wintertime could coincide with a reduction in worm populations. Temperature flux beyond the thermal (optimal) range can adversely affect the photosynthetic output of S. roscoffensis. Under laboratory conditions, oxygen production decreased by > 50% when temperature was raised from 14oC to 30oC (Thomas et al. 2023a). Data from the field site suggests that populations of S. roscoffensis survive the seasonal variation in conditions at the site, reflecting broad environmental plasticity (Thomas et al. 2023a). Intraspecific variation in temperature tolerance between the different geographical populations of S. roscoffensis is likely.
4.2 Molecular and phylogenetic assessments of S. roscoffensis and their algal symbionts
Using the cox1 mitochondrial gene, both maximum likelihood and Bayesian phylogenetic reconstructions placed all the S. roscoffensis samples together. There were some subtle differences between the worm populations tested here (Fig. 3). Our data goes some way to addressing the limited molecular and biogeographical information available for S. roscoffensis. First, the worms from Portugal were the most distantly related to those from France and Guernsey, followed by Spain, although they shared a common ancestor. Second, if we speculate the worms from France were the founder population (first discovered in Roscoff), the species has developed distinct ecotypes in the north (Guernsey and Wales), and south (Spain and Portugal). Interestingly, this gradient complements the ecological conditions that populations would experience at their respective locations. For instance, worm populations in Spain and Portugal would experience on average higher mean temperatures and longer day lengths in comparison to populations further north. While the sequences retrieved from Wales, France and Guernsey formed a subclade within the species (Fig. 3), they further placed the Welsh worms and a 10-year-old sequence from Roscoff together, whereas the two sequences we retrieved from Roscoff and Guernsey in this study were clustered. The sequences that we generated from worms taken from the Roscoff site did not show an identical match to those already in GenBank. Given the fact that the sequences in the data base are > 10 years apart, we suspect that these differences were due to single nucleotide polymorphisms (SNPs) within our sequences. Transitions between A/Gs and C/Ts occur regularly in such populations. The substitution of methionine to isoleucine is considered a “safe” substitution (Supp. Figure 1A and 1B) and does not result in a conformational change in protein structure (Bordo and Argos 1994; Ohmura et al. 2001); therefore, it is likely to persist in the population. Non-deleterious SNPs are also known to accumulate in populations that have little to no gene flow between populations, acting as a driver for natural selection, such as in the case of the worm populations (Ferchaud et al. 2020). Considering the disparate geographical areas of the worms’ known distribution, it is probable that some populations exist but have yet to be discovered. The worms have very limited mobility and are unlikely to swim across large distances. Isolation by distance also occurs to even larger, more mobile marine species such as reef fish and invertebrates due to restriction by physical barriers such as ocean currents (Planes and Fauvelot 2002; Johannesson et al. 2010). In other marine invertebrates that are isolated by distance, the ecotypes that form become locally adapted to conditions, for instance, temperature or salinity (Johannesson and André 2006; Barrett and Schluter 2008). This may also be the case for S. roscoffensis given the fact that different populations span large geographical and environmental gradients and as such, each S. roscoffensis population would be adapted to local conditions and vary in their tolerances. Of course, further field data is needed to attribute local environmental conditions to local adaptations for known populations of S. roscoffensis.
Concerning the algal symbiont, the results are more straightforward. T. convolutae is specific to all worms across all locations tested (Fig. 4). While S. roscoffensis in a laboratory setting can be manipulated to expel and switch its algal symbiont (Dupont et al. 2012; Arboleda et al. 2018; Thomas et al. 2023b), in the field we did not find any evidence that supports a more diverse symbiont profile other than T. convolutae. Our findings contradict Mettam (1979) and Mcfarlane (1982), who both claimed that populations at the Welsh field site differed in their resident algae. It should be noted that Mettam (1979) and Mcfarlane (1982) relied on microscopy, whereas our 18S rRNA data provided arguably more reliable algal species identity. While the worms do not acquire the algae directly from the parents, we have data to suggest that aposymbiotic worms can detect algae within their surrounding environment and move towards it (unpublished observations by the authors). Given the fact that the worms will reject any alga in the presence of T. convolutae, populations are likely to maintain the same symbiont species across multiple generations (Provasoli et al. 1968).
Tetraselmis convolutae CCAP66/36 and our own short-term culture maintained at Swansea University clustered together with diverse Tetraselmis spp. This difference may be due to the fact that algae, when in symbiosis, have slower growth than its free-living counterpart. For instance, algae in symbiosis has a doubling time of between 70 and 100 days, while free-living algae can double every 3 days (Wooldridge 2010). Therefore, T. convolutae may have a lower growth rate when residing inside the worm and differences seen within our trees may be due to non-deleterious SNPs. Over time, it is also possible that the cultures became contaminated.
5 Conclusion
Environmental conditions at the Welsh field site coincided with fluctuating S. roscoffensis population size, with temperature identified as the main predictor. Representative worms from the disparate populations studied here are distinct ecotypes (or species subtype). Future experiments should look to examine whether the location-specific ecotypes of S. roscoffensis differ from each other in their physiology, behaviour and other traits (local adaptations). The algal symbiont, T. convolutae, showed little genetic diversity between the worms sampled, illustrating the intimate relationship between the worms and its symbiont across many generations and locations.
Data Availability
All DNA sequences have been deposited in GenBank under the following accession numbers: OQ536360-OQ536363 (worm cox1) and OQ538146- OQ538151 (algal 18S rRNA).
References
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Androuin T, Six C, Bordeyne F et al (2020) Better off alone? New insights in the symbiotic relationship between the flatworm Symsagittifera roscoffensis and the microalgae Tetraselmis convolutae. Symbiosis 81:161–171. https://doi.org/10.1007/s13199-020-00691-y
Arboleda E, Hartenstein V, Martinez P et al (2018) An emerging system to study photosymbiosis, brain regeneration, chronobiology, and behavior: the marine acoel Symsagittifera roscoffensis. BioEssays 40:1800107
Bailly X, Laguerre L, Correc G et al (2014) The chimerical and multifaceted marine acoel Symsagittifera roscoffensis: from photosymbiosis to brain regeneration. Front Microbiol 5:498
Barrett RDH, Schluter D (2008) Adaptation from standing genetic variation. Trends Ecol Evol 23:38–44. https://doi.org/10.1016/j.tree.2007.09.008
Bordo D, Argos P (1994) The role of side-chain hydrogen bonds in the formation and stabilization of secondary structure in soluble proteins. J Mol Biol 243:504–519
Carvalho LF, Rocha C, Fleming A et al (2013) Interception of nutrient rich submarine groundwater discharge seepage on European temperate beaches by the acoel flatworm, Symsagittifera roscoffensis. Mar Pollut Bull 75:150–156. https://doi.org/10.1016/j.marpolbul.2013.07.045
Douglas AE (1983) Establishment of the symbiosis in Convoluta Roscoffensis. J Mar Biol Assoc United Kingdom 63:419–434. https://doi.org/10.1017/S0025315400070776
Douglas AE (1985) Growth and reproduction of convoluta roscoffensis containing different naturally occurring algal symbionts. J Mar Biol Assoc United Kingdom 65:871–879. https://doi.org/10.1017/S0025315400019378
Douglas AE, Gooday GW (1982) The behaviour of algal cells towards egg capsules of convoluta roscoffensis and its role in the persistence of the convoluta-alga symbiosis. Br Phycol J 17:383–388. https://doi.org/10.1080/00071618200650391
Dupont S, Moya A, Bailly X (2012) Stable photosymbiotic relationship under CO2-Induced Acidification in the Acoel Worm Symsagittifera Roscoffensis. PLoS ONE 7:e29568
Ferchaud A, Leitwein M, Laporte M et al (2020) Adaptive and maladaptive genetic diversity in small populations: insights from the Brook Charr (Salvelinus fontinalis) case study. Mol Ecol 29:3429–3445. https://doi.org/10.1111/mec.15566
Franks NR, Worley A, Grant KAJ et al (2016) Social behaviour and collective motion in plant-animal worms. Proc R Soc B Biol Sci 283:20152946
Geddes P (1879) II. Observations on the physiology and histology of Convoluta schultzii. Proc R Soc London 28:449–457
Guillard RR, Ryther JH (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can J Microbiol 8:229–239. https://doi.org/10.1139/m62-029
ITIS (2019) ITIS Standard Report Page: Symsagittifera roscoffensis. https://www.itis.gov/servlet/SingleRpt/SingleRpt#null. Accessed 18 Oct 2019
Johannesson K, André C (2006) Invited review: life on the margin: genetic isolation and diversity loss in a peripheral marine ecosystem. The Baltic Sea Mol Ecol 15:2013–2029. https://doi.org/10.1111/j.1365-294X.2006.02919.x
Johannesson K, Panova M, Kemppainen P et al (2010) Repeated evolution of reproductive isolation in a marine snail: unveiling mechanisms of speciation. Philos Trans R Soc B Biol Sci 365:1735–1747. https://doi.org/10.1098/rstb.2009.0256
Jondelius U, Wallberg A, Hooge M, Raikova OI (2011) How the worm got its pharynx: phylogeny, classification and bayesian Assessment of Character Evolution in Acoela. Syst Biol 60:845–871. https://doi.org/10.1093/sysbio/syr073
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. https://doi.org/10.1093/nar/gkz239
Lim DKY, Garg S, Timmins M et al (2012) Isolation and evaluation of oil-producing Microalgae from Subtropical Coastal and Brackish Waters. PLoS ONE 7:e40751
Martinez P, Ustyantsev K, Biryukov M, Mouton S, Glasenburg L, Sprecher SG, Bailly X, Berezikov E (2023) Genome assembly of the acoel flatworm Symsagittifera roscoffensis, a model for research on body plan evolution and photosymbiosis. G3 13(2):jkac336. https://doi.org/10.1093/g3journal/jkac336
Mcfarlane AE (1982) Two species of Algal Symbiont in naturally occurring populations of Convoluta Roscoffensis. J Mar Biol Assoc United Kingdom 62:235. https://doi.org/
Mettam C (1979) A Northern Outpost of Convoluta Roscoffensis in South Wales. J Mar Biol Assoc United Kingdom 59:251–252. https://doi.org/10.1017/S0025315400046324
Ohmura T, Ueda T, Hashimoto Y, Imoto T (2001) Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme. Protein Eng Des Sel 14:421–425. https://doi.org/10.1093/protein/14.6.421
Oschman JL (1966) Development of the symbiosis of < i > Convoluta roscoffensis < i > graff and platymonas sp. 1. J Phycol 2:105–111
Planes S, Fauvelot C (2002) Isolation by distance and vicariance drive genetic structure of a coral reef fish in the pacific ocean. Evol (N Y) 56:378–399. https://doi.org/10.1111/j.0014-3820.2002.tb01348.x
Provasoli L, Yamasu T, Manton I (1968) Experiments on the resynthesis of symbiosis in Convoluta roscoffensis with different flagellate cultures. J Mar Biol Assoc United Kingdom 48:465–478. https://doi.org/10.1017/S0025315400034603
Riewluang S, Wakeman KC (2023) Biodiversity of symbiotic microalgae associated with meiofaunal marine acoels in Southern Japan. PeerJ 11:e16078. https://doi.org/10.7717/peerj.16078
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol 9:678–687
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Thomas NJ, Coates CJ, Tang KW (2023a) Environmental constraints on the photosynthetic rate of the marine flatworm Symsagittifera roscoffensis. J Exp Mar Bio Ecol 558:151830. https://doi.org/10.1016/j.jembe.2022.151830
Thomas NJ, Tang KW, Coates CJ (2023b) Prospecting the photosynthetic flatworm Symsagittifera roscoffensis as a novel fish-feed. Aquac J 3:149–167. https://doi.org/10.3390/aquacj3020013
von Graff L (1891) Die Organisation Der Turbellaria Acoela. W. Engelmann
Wooldridge SA (2010) Is the coral-algae symbiosis really ‘mutually beneficial’ for the partners? BioEssays. 32:615–625. https://doi.org/10.1002/bies.200900182
World Sea Temperatures (2023) World Sea Temperatures. https://www.seatemperature.org/europe/united-kingdom/hull.htm. Accessed 26 Oct 2023
Acknowledgements
We would like to thank everyone who kindly provided samples from the various field locations: Mel Broadhurst-Allen (Alderney wildlife trust, Guernsey), Luis Oliveira (Portuguese Institute for Sea and Atmosphere-IMPA, Portugal), and Xavier Bailly (Station Biologique De Roscoff, France). We are grateful to Dr Jessica Bevan (née Thomas) for the technical support.
Funding
The project was funded in part by HEFCW & Swansea University RWIF (RIG1036-125) awarded to CJC and KWT.
Author information
Authors and Affiliations
Contributions
Nathan J. Thomas: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. Kam W. Tang: Conceptualization, Methodology, Resources, Writing – original draft, Writing – review & editing, Supervision, Project administration. Christopher J. Coates: Conceptualization, Methodology, Formal analysis, Resources, Writing – original draft, Writing – review & editing, Supervision, Project administration.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Thomas, N.J., Tang, K.W. & Coates, C.J. In situ environmental conditions and molecular identification of the photosymbiotic marine worm Symsagittifera roscoffensis. Symbiosis 92, 137–148 (2024). https://doi.org/10.1007/s13199-023-00964-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-023-00964-2