Abstract
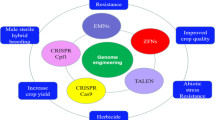
Genome editing technologies using customizable sequence specific nucleases (SSNs) enables precise genome modification in plants, which has revolutionized functional genomics research as well as crop improvement. Applicability of SSN-mediated genome editing ranges from targeted gene knockout and single base modification to multiple gene stacking into desired genomic sites. However, there are still considerable challenges in implementing genome editing technologies for practical crop improvement. Here, we briefly discuss the technological challenges, especially those associated with the delivery of SSNs and with homologous recombination-mediated genome editing, and address some promising solutions to overcome the issues.
Similar content being viewed by others
References
Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP Citovsky V, Conrad L, Gelvin SB, Jackson D, Kausch AP, Lemaux PG, Medford JI, Orozo-Cardenas M, Tricoli D, VanEck J, Voytas DF, Walbot V, Wang K, Zhang ZJ, Stewart CN. 2016. Advancing crop transformation in the era of genome editing. Plant Cell 28(7): 1510–1520
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. 2014. DNA replicons for plant genome engineering. Plant Cell 26(1): 151–163
Bartlett JG, Alves SC, Smedley M, Snape JW, Harwood WA. 2008. High-throughput Agrobacterium-mediated barley transformation. Plant Methods 4(1): 22
Bastaki NK, Cullis CA. 2014. Floral-Dip Transformation of flax (Linum usitatissimum) to generate transgenic progenies with a high transformation rate. JoVE 94: 52189
Bortesi L, Fischer R. 2015. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33(1): 41–52
Cermák T, Baltes NJ, Cegan R, Zhang Y, Voytas DF. 2015. High-frequency, precise modification of the tomato genome. Genome Biol. 16(1): 232
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186(2): 757–761
Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16(6): 735–743
Curtis IS, Nam HG. 2001. Transgenic radish (Raphanus sativusL.longipinnatus Bailey) by floral-dip method–plant development and surfactant are important in optimizing transformation efficiency. Transgenic Res. 10(4): 363–371
Ding Y, Li H, Chen L-L, Xie K. 2016. Recent advances in genome editing using CRISPR/Cas9. Front. Plant Sci. 7: 703
Elmayan T, Vaucheret H. 1996. Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9(6): 787–797
Endo M, Mikami M, Toki S. 2016. Biallelic gene targeting in rice. Plant Physiol. 170(2): 667–677
Even-Faitelson L, Samach A, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. 2011. Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J. 68(5): 929–937
Hilscher J, Bürstmayr H, Stoger E. 2016. Targeted modification of plant genomes for precision crop breeding. Biotechnol. J. 12: 1600173
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096): 816–821
Kaeppler SM, Kaeppler HF, Rhee Y. 2000. Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 43(2): 179–188
Kim YG, Cha J, Chandrasegaran S. 1996. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 93(3): 1156–1160
Kwon Y-I, Abe K, Osakabe K, Endo M, Nishizawa-Yokoi A, Saika H, Shimada H, Toki S. 2012. Overexpression of OsRecQl4 and/or OsExo1 enhances DSB-induced homologous recombination in rice. Plant Cell Physiol. 53(12): 2142–2152
Li J, Tan X, Zhu F, Guo J. 2010. A rapid and simple method for Brassica napus floral-dip transformation and selection of transgenic plantlets. Int. J. Biol. 2(1): 127–131
Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C. 2017. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nature Comm. 8: 14261
Lu C, Kang J. 2008. Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep. 27(2): 273–278
Luo S, Li J, Stoddard Thomas J, Baltes Nicholas J, Demorest Zachary L, Clasen Benjamin M, Coffman A, Retterath A, Mathis L, Voytas Daniel F, Zhang F. 2015. Non-transgenic plant genome editing using purified sequence-specific nucleases. Mol. Plant 8(9): 1425–1427
Malyska A, Bolla R, Twardowski T. 2016. The role of public opinion in shaping trajectories of agricultural biotechnology. Trends Biotechnol. 34(7): 530–534
Martin-Ortigosa S, Peterson DJ, Valenstein JS, Lin VS-Y, Trewyn BG, Lyznik LA, Wang K. 2014. Mesoporous silica nanoparticle-mediated intracellular Cre protein delivery for maize genome editing via loxP Site excision. Plant Physiol. 164(2): 537–547
Martin-Ortigosa S, Wang K. 2014. Proteolistics: a biolistic method for intracellular delivery of proteins. Transgenic Res. 23(5): 743–756
Martins PK, Nakayama TJ, Ribeiro AP, Cunha BADBd, Nepomuceno AL, Harmon FG, Kobayashi AK, Molinari HBC. 2015. Setaria viridis floral-dip: A simple and rapid Agrobacterium-mediated transformation method. Biotechnol. Rep. 6: 61–63
Matzke AJM, Matzke MA. 1998. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1(2): 142–148
Morbitzer R, Römer P, Boch J, Lahaye T. 2010. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl. Acad. Sci. USA 107(50): 21617–21622
Mu G, Chang N, Xiang K, Sheng Y, Zhang Z, Pan G. 2012. Genetic transformation of maize female inflorescence following floral dip method mediated by Agrobacterium. Biotechnol. Adv. 11(3): 178–183
Nishizawa-Yokoi A, Nonaka S, Osakabe K, Saika H, Toki S. 2015. A universal positive-negative selection system for gene targeting in plants combining an antibiotic resistance gene and its antisense RNA. Plant Physiol. 169(1): 362–370
Pellegrineschi A, Noguera LM, Skovmand B, Brito RM, Velazquez L, Salgado MM, Hernandez R, Warburton M, Hoisington D. 2002. Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome 45(2): 421–430
Pitzschke A, Hirt H. 2010. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 29(6): 1021–1032
Popat A, Hartono SB, Stahr F, Liu J, Qiao SZ, Qing Lu G. 2011. Mesoporous silica nanoparticles for bioadsorption, enzyme immobilisation, and delivery carriers. Nanoscale 3(7): 2801–2818
Puchta H. 2005. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 56(409): 1–14
Puchta H. 2017. Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr. Opin. Plant Biol. 36: 1–8
Reiss B, Schubert I, Köpchen K, Wendeler E, Schell J, Puchta H. 2000. RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc. Natl. Acad. Sci. USA 97(7): 3358–3363
Rod-in W, Sujipuli K, Ratanasut K. 2014. The floral-dip method for rice (Oryza sativa) transformation. J. Agri. Technol. 10(2): 467–474
Sarmast MK. 2016. Genetic transformation and somaclonal variation in conifers. Plant Biotechnol. Rep. 10(6): 309–325
Sauer NJ, Narváez-Vásquez J, Mozoruk J, Miller RB, Warburg ZJ Woodward MJ, Mihiret YA, Lincoln TA, Segami RE, Sanders SL, Walker KA, Beetham PR, Schöpke CR, Gocal GF. 2016. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 170(4): 1917–1928
Shimatani Z, Nishizawa-Yokoi A, Endo M, Toki S, Terada R. 2015. Positive–negative-selection-mediated gene targeting in rice. Front. Plant Sci. 5: 748
Smith J, Grizot S, Arnould S, Duclert A, Epinat J-C Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, Montoya G, Pâques F, Duchateau P. 2006. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 34(22): e149
Steinert J, Schiml S, Puchta H. 2016. Homology-based doublestrand break-induced genome engineering in plants. Plant Cell Rep. 35(7): 1429–1438
Stoddard TJ, Clasen BM, Baltes NJ, Demorest ZL, Voytas DF, Zhang F, Luo S. 2016. Targeted mutagenesis in plant cells through transformation of sequence-specific nuclease mRNA. PLOS One 5: e0154634
Subburaj S, Chung SJ, Lee C, Ryu S-M, Kim DH, Kim J-S, Bae S, Lee G-J. 2016. Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35(7): 1535–1544
Sun Y, Li J, Xia L. 2016a. Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front. Plant Sci. 7: 1928
Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L. 2016b. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 9(4): 628–631
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. 2015. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169(2): 931–945
Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S. 2002. Efficient gene targeting by homologous recombination in rice. Nat. Biotech. 20(10): 1030–1034
Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. 2009. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459(7245): 442–445
Trieu AT, Burleigh SH, Kardailsky IV, Maldonado-Mendoza IE, Versaw WK Blaylock LA, Shin H, Chiou TJ, Katagi H, Dewbre GR, Weigel D, Harrison MJ. 2000. Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 22(6): 531–541
Voytas DF. 2013. Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64(1): 327–350
Waterworth WM, Drury GE, Bray CM, West CE. 2011. Repairing breaks in the plant genome: the importance of keeping it together. New Phytol. 192(4): 805–822
Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim S-G, Kim S-T, Choe S, Kim J-S. 2015. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotech. 33(11): 1162–1164
Wright DA, Townsend JA, Winfrey RJ, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. 2005. Highfrequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 44(4): 693–705
Zale JM, Agarwal S, Loar S, Steber CM. 2009. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Rep. 28(6): 903–913
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jung, J.H., Seo, Y.W. Challenges in wide implementation of genome editing for crop improvement. J. Crop Sci. Biotechnol. 20, 129–135 (2017). https://doi.org/10.1007/s12892-017-0019-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-017-0019-0