Abstract
We used double digest restriction site associated DNA sequencing (ddRAD) to develop new geographically informative nuclear SNP loci in Quercus robur and Quercus petraea. Genotypes derived from sequence data of 95 individuals covering the distribution range of the species were analysed to select geographically informative and polymorphic loci within Russia and Germany. We successfully screened a selected set of 119 loci on a MassARRAY® iPLEX™ platform on 190 individuals from 19 locations in Russia. The newly developed loci will be useful for genetic studies over the whole distribution range of both species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Quercus robur (L.) and Quercus petraea (Matt.) are very common tree species in many forests in Europe. Their distribution range from Scandinavia to the Iberian Peninsula, and from Great Britain towards Eastern and South-Eastern Europe (San-Miguel-Ayanz et al. 2016). The distribution edge of Quercus robur goes a few hundred kilometres more to the East up to the South-Ural region in Russia. Both species are economically significant and provide high quality hardwood for timber construction, furniture and barrels.
Quercus robur and Q. petraea have been genetically very well studied over their West-European distribution range, but there are still some gaps especially to the East (Degen et al. 2019). As gene markers, chloroplastic RFLPs, microsatellites and recently SNPs have been applied to study genetic differentiation and population genetic processes like gene flow, mating system and the impact of post glacial re-colonisation and human seed transfer (Konig et al. 2002; Petit et al. 2002; Buschbom et al. 2011; Gerber et al. 2014). Although large sets of SNPs including thousand to millions of SNPs have been developed for the two oak species (Leroy et al. 2019; Lepoittevin et al. 2015), there is still a need for smaller sets of highly geographically informative SNPs that can be cost-effectively genotyped.
For SNP discovery, we used leaf or cambium material from 95 Q. robur and Q. petraea trees originating from all Europe, Ukraine and Russia (Table 1). The sampling size was more intensive in Germany to catch within-country variation. DNA was extracted according to Dumolin et al. (1995). Double Digest Restriction site associated DNA sequencing (ddRAD) (Peterson et al. 2012) was conducted on all samples to detect SNPs in the nuclear genome (Floragenex, Portland, USA). Among the 26,074 putative SNP loci obtained by ddRADseq, only those with a minimum flanking region of 50 bp around the SNP and a maximum of two neighbour SNPs were selected for further analysis (3648 loci). Further cleaning of the data included the removal of loci with more than 10% missingness and a minor allele frequency lower than 1%. Data was grouped by species, country, and state within Germany to conduct discriminant analysis and to detect the loci with the highest contribution. This allowed a selection of SNP loci with a geographical signal. We additionally looked among samples from Germany and Russia to select loci with both high expected heterozygosity and positive Fis to avoid parapatric loci. All analyses were conducted in R 3.6.0 using, among others, the packages vcfR, poppR, adegenet and hierfstat. A final selection of 168 loci was used to design four MassARRAY ® iPLEX™ multiplexes (Assay Design Suite v2.0 [Agena Bioscience™, San Diego, USA]), which included a total of 130 loci (Supplementary Material S1).
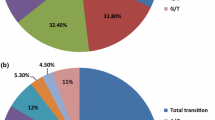
We choose to test our newly developed markers on 190 Quercus robur from 19 locations in its Russian distribution range (Table 2). All samples were run on a MassARRAY® iPLEX™ platform (Agena Bioscience™, San Diego, USA) using the iPLEX™ GOLD chemistry. Genotyping was conducted with Typer Viewer v.4.0.24.71 (Agena Bioscience™, San Diego, USA). We estimated for each locus the percentage of amplification, observed heterozygosity (Ho), within population gene diversity (Hs) (Nei 1987), Fis, Fst (Weir and Cockerham 1984) and average differentiation among sampling locations (Gregorius 1987). Significance levels for Fis and Fst were tested with 10,000 randomizations (Supplementary Material S2). Analyses were conducted with GDA_NT (Degen, unpublished) and Fstat (Goudet 1995). A total of 119 markers was usable. Among those, four loci showed an amplification lower than 80% and 24 loci were not polymorphic in the screened Russian samples. Additionally, nine loci significantly deviated from HWE in the tested samples. Differentiation was low (Fst = 0.03) but significant (Goudet et al. 1996).
We successfully developed new SNP markers for Q. robur and Q. petraea, which will be useful for population genetic studies at the European level. Additional screening will be needed on samples from other regions, to address whether a single set of SNP markers will be sufficient to cover the whole distribution range of these species and be useful to track the origin of forest reproductive material.
Change history
30 June 2021
A Correction to this paper has been published: https://doi.org/10.1007/s12686-021-01222-7
References
Buschbom J, Yanbaev Y, Degen B (2011) Efficient long-distance gene flow into an isolated relict oak stand. J Hered 102:464–472
Degen B, Yanbaev R, Yanbaev Y (2019) Genetic differentiation of Quercus robur in the South-Ural. Silvae Genet 68:111–115
Dumolin S, Demesure B, Petit RJ (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor Appl Genet 91:1253–1256
Gerber S, Chadoeuf J, Gugerli F, Lascoux M, Buiteveld J, Cottrell J, Dounavi A, Fineschi S, Forrest LL, Fogelqvist J, Goicoechea PG, Jensen JS, Salvini D, Vendramin GG, Kremer A (2014) High rates of gene flow by pollen and seed in oak populations across Europe. PLoS ONE 9:16
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J, Raymond M, deMeeus T, Rousset F (1996) Testing differentiation in diploid populations. Genetics 144:1933–1940
Gregorius HR (1987) The relationship between the concepts of genetic diversity and differentiation. Theor Appl Genet 74:397–401
Hartl DL, Clark AG (1997) Principles of population genetics. Sinauer Associates, Sunderland MA
Konig AO, Ziegenhagen B, van Dam BC, Csaikl UM, Coart E, Degen B, Burg K, de Vries SMG, Petit RJ (2002) Chloroplast DNA variation of oaks in western Central Europe and genetic consequences of human influences. For Ecol Manag 156:147–166
Lepoittevin C, Bodenes C, Chancerel E, Villate L, Lang T, Lesur I, Boury C, Ehrenmann F, Zelenica D, Boland A, Besse C, Garnier-Gere P, Plomion C, Kremer A (2015) Single-nucleotide polymorphism discovery and validation in high-density SNP array for genetic analysis in European white oaks. Mol Ecol Res 15:1446–1459
Leroy T, Rougemont Q, Dupouey JL, Bodenes C, Lalanne C, Belser C, Labadie K, Le Provost G, Aury JM, Kremer A, Plomion C (2019) Massive postglacial gene flow between European white oaks uncovered genes underlying species barriers. New Phytol. https://doi.org/10.1111/nph.16039
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135
Petit RJ et al (2002) Chloroplast DNA variation in European white oaks—phylogeography and patterns of diversity based on data from over 2600 populations. For Ecol Manag 156:5–26
San-Miguel-Ayanz J, De Rigo D, Caudullo G, Durrant TH, Mauri A (2016) European atlas of forest tree species. Publications Office of the European Union
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population-structure. Evolution 38:1358–1370
Acknowledgements
The study was co-financed by Grant WKF-WF04-22WC4111 01 (German Federal Ministry of Food and Agriculture & German Federal German Ministry for the Environment, Nature Conservation and Nuclear Safety) and for the study on the Russian pedunculate oak samples by the Grant No 19-16-00084 from the Russian Science Foundation (project members: S. Bakhtina, R. Yanbaev, Y. Yanbaev and B. Degen). Genotyping was performed at the Genome Transcriptome Facility of Bordeaux (Grants from ANR-10-EQPX-16), with the help of Marie Massot. We are grateful to Stefan Jencsik for his work with sampling in provenance trial and with laboratory work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised due to retrospective open access order.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Blanc-Jolivet, C., Bakhtina, S., Yanbaev, R. et al. Development of new SNPs loci on Quercus robur and Quercus petraea for genetic studies covering the whole species’ distribution range. Conservation Genet Resour 12, 597–600 (2020). https://doi.org/10.1007/s12686-020-01141-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-020-01141-z