Abstract
Better-informed mangrove conservation and management practices are needed as the ecosystem services provided by these intertidal forests continue to be threatened by increasing anthropogenic pressures and climate change. Multiple layers of knowledge are required to achieve this goal, including insights into population genetics of mangrove species. Understanding the importance of population-genetic insights to conservation, multiple research groups have developed microsatellite loci for the widespread, neotropical red mangrove, Rhizophora mangle. However, although a wealth of genetic markers exist, empirical research is limited in the number of these loci employed. Here, we designed two multiplex PCR panels that combine seven novel loci developed for this work and eight previously-developed loci from three research groups to generate 15-locus genotypes, more than twice the average number of loci used in previous research, in only two PCR. We demonstrated utility in R. mangle from four sites across ~ 2500 km near this species’ northern latitudinal limits, and that these multiplex panels were better able to delineate populations than data subsets with numbers of loci comparable to previous research. We focus our discussion on how this tool is a more-informative, efficient (both in terms of time and resources), and easily-modifiable alternative to address many pressing conservation and management issues, such as the generation of baseline genetic data for areas not yet studied, better defining management units, and monitoring genetic effects of restoration projects. We also provide a quick protocol that outlines each step in this procedure to facilitate the use of this tool by others.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mangroves provide ecosystem services of both ecological and economic importance to coastal ecosystems worldwide (Lee et al. 2014). However, these intertidal forests are highly susceptible to increasing anthropogenic pressures and climate change (Alongi 2015; Friess et al. 2019). Effective conservation and management of mangrove ecosystems will require multiple layers of knowledge across diverse disciplines, including improved estimates of temporal changes in mangrove cover, standardized protocols to monitor forests, and insights into connectivity across local and regional scales (Canty et al. 2018). Population genetic data can provide insights necessary to understand and continue to monitor species for conservation and management purposes, including estimates of population structure, effective population sizes, and gene flow (Kramer and Havens 2009).
The pantropical genus Rhizophora (Rhizophoraceae) consists of nine species and hybrids (Duke et al. 1998) whose large propagules are commonly used in reforestation projects. Three members of this genus exist in the Neotropics, where Rhizophora mangle is the most widespread, with a distribution that covers both the Pacific and Atlantic coasts of the Americas and the Atlantic coast of Africa. Understanding the importance of population-genetic insights to mangrove conservation, multiple research groups have developed R. mangle microsatellite loci (Rosero‐Galindo et al. 2002; Takayama et al. 2008; Ribeiro et al. 2013; Francisco et al. 2018a) and 17 peer-reviewed publications since 2007 have utilized these loci to characterize R. mangle population genetics from across this species’ distribution (Table 1). However, although we possess a wealth of genetic markers, this field still lacks cohesion in the implementation of these microsatellite loci. Empirical research is limited in the number of these loci employed (6.6 ± 2 loci; mean ± SD), presumably because most studies continue to amplify loci individually (Table 1). Amplification in singleplex can be excessively expensive and time consuming, and limit either the number of markers used or samples genotyped, as expressed in a recent study (Bologna et al. 2019). In addition, many studies limit themselves to loci developed by a single research group and do not use previous research to inform their choice of loci from across all available microsatellites.
Here, we developed a new set of R. mangle microsatellite loci and then designed two multiplex PCR panels that combine these novel loci with those of three other research groups. These multiplex panels generate 15-locus genotypes, more than twice the average number of loci used in previous research, in only two PCR. We demonstrate the utility of these multiplex panels in R. mangle from four collection sites across ~ 2500 km towards this species’ northern latitudinal limits and how this increased number of loci can improve our ability to differentiate among populations of this species. We focus our discussion on how this tool can be an efficient alternative (both in terms of time and resources) to provide necessary baseline genetic data for pressing conservation and management questions, and how these multiplex can be easily modified to incorporate alternative loci from the pool of available microsatellites for this species.
Materials and methods
Novel microsatellites
Rhizophora mangle leaf tissue was collected from a single individual in Fort Pierce, Florida, USA (27.4974, − 80.3057) and immediately dried in silica gel. Genomic DNA from this individual was isolated from 20 mg of dried leaf tissue with the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) according to manufacturer’s protocol, with an extended incubation of 45 min. DNA for sequencing was purified with the High Pure PCR Product Purification Kit (Roche, Penzberg, Germany). We used 2 × 250-bp paired-end Illumina MiSeq genome sequencing and developed microsatellite markers with the Galaxy-based pipeline outlined by Griffiths et al. (2016), which identified 61,130 sequences with microsatellite motifs and designed primers for 358 loci, of which 42 had assembled read sequences. A subset of 34 of these loci was selected from across all possible perfect repeat motifs (di-, tri-, tetra-, and pentanucleotides) and tested with 16 R. mangle individuals from two collection sites in Florida, USA (Avalon: 27.5468, -80.3297; Pine Island: 28.4841, − 80.7237; n = 8 per site). We used the DNeasy Plant Mini Kit to isolate genomic DNA from these 16 individuals, as described above.
We aimed to combine multiple loci into a limited number of multiplex reactions, so we performed singleplex testing for all loci with identical PCR conditions. We followed the PCR method for a single set of cycles outlined in Culley et al. (2013): 95 °C for 15 min; 35 cycles at 94 °C for 30 s, 57 °C for 90 s, 72 °C for 60 s; 60 °C for 30 min. We used the Type-it® Microsatellite PCR Kit (Qiagen, Valencia, California, USA) with a total volume of 6 μL per reaction, with 2.5 μL Multiplex PCR Master Mix, 0.5 μL primer mix (0.2 μM of each forward and reverse), 1 μL dH2O, and 2 μL of genomic DNA (~ 20 ng). We performed PCR on a Prime thermal cycler (Techne, Straffordshire, UK), and assessed amplification via electrophoresis on 1.5% agarose gels. Of the 34 loci, 18 produced consistent bands, whereas others did not amplify or produced multiple size bands. For these 18 loci, we ordered new forward primers with additional sequences at the 5′ end that correspond to universal primers with fluorescent labels (6-FAM, HEX, or PET), and used a three-primer method to fluorescently label PCR products, as described in Culley et al. (2013). We analysed fragments on an Applied Biosystems 3730 DNA Analyzer (Applied Biosystems, Foster City, California, USA) and scored alleles in the R-package Fragman (Covarrubias-Pazaran et al. 2016). A total of 15 loci produced easily-identifiable peaks and nine were polymorphic in the 16 Florida samples (Table 2).
Multiplex design
Utilizing the same PCR conditions and three-primer method outlined above, we initiated multiplex testing with 28 loci (15 developed here and 13 published). We selected RM19, RM38, RM41 (Rosero‐Galindo et al. 2002), and RM50, RM86 (Takayama et al. 2008) based on our previous experience (Kennedy et al. 2016, 2017), and selected eight loci developed by Ribeiro et al. (2013): RmBra18, RmBra19, RmBra20, RmBra25, RmBra50, RmBra59, RmBra64, RmBra66. RmBra25 was discarded due to inconsistent singleplex amplification. We combined the remaining 27 loci into three initial multiplexes based simply on fragment length differences. Loci were discarded due to inconsistent multiplex amplification (RzMg07, RzMg30, RM86), difficult-to-score peaks (RmBra66, RM41) or monomorphism (RzMg04, RzMg05, RzMg08, RzMg16, RzMg18, RzMg25, RmBra64).
We combined the remaining 15 loci into two multiplex reactions with seven and eight loci each (Table 3). We used the same PCR volumes and conditions described above (see Appendix S1 for a protocol outline).
Multiplex testing
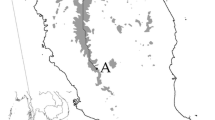
We assessed multiplexes with 103 R. mangle individuals from four collection sites: one site in Florida, USA with 31 samples (Jupiter: 26.8179, − 80.0480), two sites in The Bahamas, at either end of the archipelago, with 35 samples (New Providence: 24.9920, − 77.3868) and 33 samples (Inagua: 21.0954, − 73.6300), and one site at this species’ northern limit in Texas, USA where only four trees were found (Río Bravo: 25.9526, − 97.1513) (Fig. 1). Distances between collection sites range from approximately 335 km (Jupiter–New Providence) to 2500 km (Río Bravo–Inagua). We used the DNeasy Plant Mini Kit to isolate genomic DNA from these individuals, as described above, and voucher material from each collection site was deposited at the Manchester Museum Herbarium (Table 4). We analysed PCR products and scored alleles as described above (see Appendix S2 for genotype data). For each collection site, we determined the number of alleles and private alleles per locus, calculated observed and expected heterozygosity, calculated inter-site genetic differentiation (FST), and tested for linkage disequilibrium and deviations from Hardy–Weinberg equilibrium after adjusting for multiple comparisons with FSTAT 2.9.3.2 (Goudet 2002). Only allele numbers were determined for the four Texas individuals.
Four Rhizophora mangle collection sites towards this species’ northern latitudinal limits. From left to right: Río Bravo, Texas (triangle), Jupiter, Florida (circle), New Providence, The Bahamas and Inagua, The Bahamas (squares). Neotropical mangrove distribution shown in green (Giri et al. 2011)
We then evaluated the ability of the 15-locus genotypes generated from these multiplexes to differentiate among the three collection sites from Florida and the Bahamas (inter-site distances ranged from 335–900 km; n = 99 individuals) compared to genotypes with fewer loci, comparable to numbers utilized in previous research (Table 1). To do this, we performed a discriminant analysis of principal components (DAPC) (Jombart et al. 2010) in the R-package adegenet 2.1.1 (Jombart and Ahmed 2011) in R v3.4.2 (R Core Team 2013). DAPC first transforms data with a principal components analysis and then performs a discriminant analysis on the principal components retained (Jombart and Collins 2015). We performed an initial analysis on the complete data set (i.e., individuals with 15-locus genotypes from both multiplexes), and then subsequent analyses on subsets with 7-locus genotypes (i.e., only data from multiplex 1) and with 8-locus genotypes (i.e., only data from multiplex 2). For each analysis, we retained the minimum number of principal components that explained ~ 90% of the total variance, which corresponded to 15, 9, and 8 principal components, respectively, and then retained both discriminant functions. We extracted each individual’s coordinates on the two principal axes of the DAPC (i.e., ind.coord) to then plot in ggplot2 (Wickham 2011).
Results
We found that 14 of the 15 loci in the two multiplexes were polymorphic across these four collection sites (Table 4). We identified 57 total alleles (Table 3), with a range from 44 alleles (Jupiter, Florida) to only 20 alleles (Río Bravo, Texas). Alleles per locus within sites ranged from 1 to 6, with expected heterozygosity from 0.00 to 0.72, and 16 private alleles were identified (Table 4). FST indicated considerable genetic differences, with a range from 0.22 (Jupiter–New Providence) to 0.52 (Jupiter–Inagua). Expected heterozygosity was generally higher than observed, but few loci deviated from Hardy–Weinberg equilibrium (Table 4). We found no evidence of linkage disequilibrium.
DAPC with the complete dataset of 15-locus genotypes (14 loci were polymorphic), identified clear delineations between the three collection sites in Florida and the Bahamas (Fig. 2A). In contrast, subsets with 7-locus and 8-locus genotypes (6 and 8 loci were polymorphic, respectively) identified a similar pattern, but were unable to clearly differentiate these collection sites (Fig. 2B, C).
Multiplex PCR panels performed better than data subsets with numbers of loci comparable to previous research. Scatterplots of discriminant analysis of principal components (DAPC) for A the complete data set (both multiplexes with 14 polymorphic loci), B only multiplex 1 (6 polymorphic loci), C only multiplex 2 (8 polymorphic loci). Individuals from Jupiter, Florida are shown with blue squares, New Providence, The Bahamas are shown with red circles, and Inagua, The Bahamas are shown with green triangles
Discussion
Better-informed mangrove conservation practices are needed as the ecosystem services provided by these intertidal forests continue to be threatened by increasing anthropogenic pressures and climate change (Friess et al. 2019). Population genetic data can provide insights necessary to understand and continue to monitor species for conservation and management purposes (Schwartz et al. 2007; Kramer and Havens 2009). Understanding the importance of population-genetic insights to mangrove conservation, researchers have made a substantial effort to develop genetic markers for the widespread neotropical red mangrove, Rhizophora mangle. There are now a total of 57 microsatellite loci available for R. mangle, with 42 previously-published loci (Rosero‐Galindo et al. 2002; Takayama et al. 2008; Ribeiro et al. 2013; Francisco et al. 2018a) and 15 novel loci from this work. Yet, empirical research, on average, employs less than seven of these available loci, likely because few studies have incorporated multiplex reactions (Table 1). Here, we outlined multiplex PCR panels that combine efforts of four geographically-distant research groups into a tool that should enable us to better outline genetic patterns in this widespread species, and do so with considerable less investment in time and resources. In this discussion, we highlight the continued utility of genetic data in mangrove conservation in the era of next-generation sequencing and urge researchers to use, modify, and improve upon this genetic tool to characterize R. mangle population genetics across the Neotropics and answer pressing conservation questions.
Conservation research seems to be in a transition from genetics to genomics as we continue to improve our ability to generate and analyse high-throughput sequence data (Puckett 2017). Genomics will enable researchers to address many new questions and, in certain contexts, provide greater resolution, but the investment in increased data is not always needed (Shafer et al. 2015). To address certain questions, and at certain spatial-scales, genetic data sets may prove sufficient and much more cost effective (Shafer et al. 2015; Puckett 2017), and this certainly seems true in terms of many outstanding questions in mangrove conservation. A reliable panel of microsatellites would be more appropriate for smaller-scale studies with moderate sample sizes, which constitutes most R. mangle research to date (Table 1), or when repeated measures are needed, as in the case of monitoring ongoing reforestation projects, as genome sequencing is most cost effective with large numbers of samples (Puckett 2017). Low quantity and quality DNA, as is often the case in mangrove species because leaf tissues are rich in molecular by-products (Huang et al. 2002), can also be an impediment to sequence library preparation, but microsatellite amplification often requires limited DNA template. Marker development is the principal investment for microsatellites, but this cost has already been paid by the multiple research groups outlined above, and many of these markers have been tested in multiple published works. The multiplex PCR panels outlined here are the product of these diverse genetic resources and discoveries, and should provide an easy-to-use and cost-effective (both in terms of time and resources) tool.
Of course, the utility of this tool relies on whether microsatellites provide sufficient polymorphism to answer conservation and management questions across the broad distribution of this species. We demonstrated that the 14 polymorphic loci in these multiplex panels, clearly delineated three populations near the northern limits of this species’ distribution and performed better than data subsets with loci numbers comparable to previous research. Although perhaps obvious, greater resolution with these multiplex panels is the result of genotyping twice as many loci as previous studies, congruent with observations based on SNP loci (Hodel et al. 2017), and of utilizing our previous experience to selectively choose loci that have proven informative. However, we have only shown that these multiplex panels are an efficient tool to genotype R. mangle from four populations across ~ 2500 km, a fraction of the entire distribution of this species. Although we are confident these multiplex panels will prove informative across a much broader spatial scale, we also envision this tool as a framework that can easily be modified depending on variation in the pool of available microsatellite loci for a particular region. For instance, Kennedy et al. (2016) discarded two loci (RM21, RM46; Rosero‐Galindo et al. 2002) due to monomorphism across much of the Caribbean, a pattern also observed in Atlantic Mexico (Cisneros-de la Cruz et al. 2018). These same loci exhibited considerable polymorphism in Pacific R. mangle from Panama, Nicaragua, and Mexico (Cerón-Souza et al. 2012; Sandoval-Castro et al. 2012, 2014; Bruschi et al. 2014). Researchers can modify these multiplex primer mixes (see Appendix S1) to include additional informative loci and/or exclude loci that exhibit monomorphism, while maintaining only two PCR per sample. This framework is much more cost effective than protocols used in previous research and should enable the inclusion of more samples and collection sites in future research. These multiplex panels should also facilitate further genetic studies to address multiple pressing conservation questions, such as generating baseline genetic data from areas that have not been studied [i.e., much of Central America, Pacific South America, Caribbean islands, and West Africa (although considerable work has been done in R. racemosa; Ngeve et al. 2016)], better defining management units to prioritize conservation measures (Wee et al. 2019), and monitoring genetic effects of restoration projects (Granado et al. 2018).
Conclusions
We developed multiplex panels with novel and published Rhizophora mangle microsatellite loci to generate 15-locus genotypes, more than twice the average number of loci used in previous research, in only two PCR (see Appendix S1 for quick protocol). We demonstrated utility across ~ 2500 km of this species’ widespread distribution, and that these multiplex panels were better able to delineate three populations near the northern limits of this species’ distribution than data subsets with numbers of loci comparable to previous research. This tool improves our ability to characterize R. mangle genetic patterns while saving researchers considerable time and resources, enables future research to include more samples and collection sites, can be easily modified to incorporate alternative informative loci, and should facilitate studies to answer multiple pressing conservation and management questions.
References
Alongi DM (2015) The impact of climate change on mangrove forests. Curr Clim Chang Rep 1:30–39. https://doi.org/10.1007/s40641-015-0002-x
Arbeláez-Cortes E, Castillo-Cárdenas MF, Toro-Perea N, Cárdenas-Henao H (2007) Genetic structure of the red mangrove (Rhizophora mangle L.) on the Colombian Pacific detected by microsatellite molecular markers. Hydrobiologia 583:321–330. https://doi.org/10.1007/s10750-007-0622-9
Bologna P, Campanella JJ, Restaino DJ, et al (2019) Lingering impacts of hurricane hugo on Rhizophora mangle (Red Mangrove) Population Genetics on St. John, USVI. Diversity 11:65. doi: 10.3390/d11040065
Bruschi P, Angeletti C, González O et al (2014) Genetic and morphological variation of Rhizophora mangle (red mangrove) along the northern Pacific coast of Nicaragua. Nord J Bot 32:320–329. https://doi.org/10.1111/j.1756-1051.2013.00138.x
Canty SWJ, Preziosi RF, Rowntree JK (2018) Dichotomy of mangrove management: a review of research and policy in the Mesoamerican reef region. Ocean Coast Manag 157:40–49. https://doi.org/10.1016/j.ocecoaman.2018.02.011
Cerón-Souza I, Bermingham E, McMillan WO, Jones FA (2012) Comparative genetic structure of two mangrove species in Caribbean and Pacific estuaries of Panama. BMC Evol Biol 12:205. https://doi.org/10.1186/1471-2148-12-205
Cerón-Souza I, Gonzalez EG, Schwarzbach AE et al (2015) Contrasting demographic history and gene flow patterns of two mangrove species on either side of the Central American Isthmus. Ecol Evol 5:3486–3499. https://doi.org/10.1002/ece3.1569
Cerón-Souza I, Rivera-Ocasio E, Medina E et al (2010) Hybridization and introgression in new world red mangroves, Rhizophora (Rhizophoraceae). Am J Bot 97:945–957. https://doi.org/10.3732/ajb.0900172
Cerón-Souza I, Turner BL, Winter K et al (2014) Reproductive phenology and physiological traits in the red mangrove hybrid complex (Rhizophora mangle and R. racemosa) across a natural gradient of nutrients and salinity. Plant Ecol 215:481–493. https://doi.org/10.1007/s11258-014-0315-1
Cisneros-de la Cruz DJ, Martínez-Castillo J, Herrera-Silveira J et al (2018) Short-distance barriers affect genetic variability of Rhizophora mangle L. in the Yucatan Peninsula. Ecol Evol 8:11083–11099. https://doi.org/10.1002/ece3.4575
Covarrubias-Pazaran G, Diaz-Garcia L, Schlautman B et al (2016) Fragman: an R package for fragment analysis. BMC Genet 17:1–8. https://doi.org/10.1186/s12863-016-0365-6
Culley TM, Stamper TI, Stokes RL et al (2013) An efficient technique for primer development and application that integrates fluorescent labeling and multiplex PCR. Appl Plant Sci 1:1300027. https://doi.org/10.3732/apps.1300027
Duke NC, Ball MC, Ellison JC (1998) Factors influencing biodiversity and distributional gradients in mangroves. Glob Ecol Biogeogr Lett 7:27–47. https://doi.org/10.2307/2997695
Francisco PM, Mori GM, Alves FM et al (2018a) Population genetic structure, introgression, and hybridization in the genus Rhizophora along the Brazilian coast. Ecol Evol 8:3491–3504. https://doi.org/10.1002/ece3.3900
Francisco PM, Tambarussi EV, de Alves FM et al (2018b) Genetic diversity and mating system of Rhizophora mangle L. (Rhizophoraceae) in Northern Brazil revealed by microsatellite analysis. CERNE 24:295–302. https://doi.org/10.1590/01047760201824042575
Friess DA, Rogers K, Lovelock CE et al (2019) The state of the world’s mangrove forests: past, present, and future. Annu Rev Environ Resour 44:89–115. https://doi.org/10.1146/annurev-environ-101718-033302
Giri C, Ochieng E, Tieszen LL et al (2011) Status and distribution of mangrove forests of the world using earth observation satellite data. Glob Ecol Biogeogr 20:154–159. https://doi.org/10.1111/j.1466-8238.2010.00584.x
Goudet J (2002) FSTAT (Version 2.9.3.2): a program to estimate and test gene diversities and fixation indices. Retrieved from http://www2.unil.ch/popgen/softwares
Granado R, Neta LCP, Nunes-Freitas AF et al (2018) Assessing genetic diversity after mangrove restoration in Brazil: why is it so important? Diversity 10:27. https://doi.org/10.3390/d10020027
Griffiths SM, Fox G, Briggs PJ et al (2016) A galaxy-based bioinformatics pipeline for optimised, streamlined microsatellite development from Illumina next-generation sequencing data. Conserv Genet Resour 8:481–486. https://doi.org/10.1007/s12686-016-0570-7
Hodel RGJ, Chen S, Payton AC et al (2017) Adding loci improves phylogeographic resolution in red mangroves despite increased missing data: comparing microsatellites and RAD-Seq and investigating loci filtering. Sci Rep 7:1–14. https://doi.org/10.1038/s41598-017-16810-7
Hodel RGJ, de Souza Cortez MB, Soltis PS, Soltis DE (2016) Comparative phylogeography of black mangroves (Avicennia germinans) and red mangroves (Rhizophora mangle) in Florida: testing the maritime discontinuity in coastal plants. Am J Bot 103:730–739. https://doi.org/10.3732/ajb.1500260
Huang Y, Shi S, Zhong Y, Tan F (2002) A new method for preparation of template DNA for PCR from special plant materials. Chinese Sci Bull 47:725–727. https://doi.org/10.1360/02tb9163
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071. https://doi.org/10.1093/bioinformatics/btr521
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jombart T, Collins C (2015) A tutorial for discriminant analysis of principal components (DAPC) using adegenet 2.0. 0. London Imp Coll London, MRC Cent Outbreak Anal Model
Kennedy JP, Garavelli L, Truelove NK et al (2017) Contrasting genetic effects of red mangrove (Rhizophora mangle L.) range expansion along West and East Florida. J Biogeogr 44:335–347. https://doi.org/10.1111/jbi.12813
Kennedy JP, Pil MW, Proffitt CE et al (2016) Postglacial expansion pathways of red mangrove, Rhizophora mangle, in the Caribbean Basin and Florida. Am J Bot 103:260–276. https://doi.org/10.3732/ajb.1500183
Kramer AT, Havens K (2009) Plant conservation genetics in a changing world. Trends Plant Sci 14:599–607. https://doi.org/10.1016/j.tplants.2009.08.005
Lee SY, Primavera JH, Dahdouh-Guebas F et al (2014) Ecological role and services of tropical mangrove ecosystems: a reassessment. Glob Ecol Biogeogr 23:726–743. https://doi.org/10.1111/geb.12155
Ngeve MN, Van der Stocken T, Menemenlis D et al (2016) Contrasting effects of historical sea level rise and contemporary ocean currents on regional gene flow of Rhizophora racemosa in Eastern Atlantic Mangroves. PLoS ONE 11:e0150950. https://doi.org/10.1371/journal.pone.0150950
Pil MW, Boeger MRT, Muschner VC et al (2011) Postglacial north-south expansion of populations of Rhizophora mangle (Rhizophoraceae) along the Brazilian coast revealed by microsatellite analysis. Am J Bot 98:1031–1039. https://doi.org/10.3732/ajb.1000392
Puckett EE (2017) Variability in total project and per sample genotyping costs under varying study designs including with microsatellites or SNPs to answer conservation genetic questions. Conserv Genet Resour 9:289–304. https://doi.org/10.1007/s12686-016-0643-7
R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. Website: R-project.org
Ribeiro DO, Vinson CC, Nascimento DSS et al (2013) Isolation of Microsatellite Markers for the Red Mangrove, Rhizophora mangle (Rhizophoraceae). Appl Plant Sci 1:1300003. https://doi.org/10.3732/apps.1300003
Rosero-Galindo C, Gaitan-Solis E, Cárdenas-Henao H et al (2002) Polymorphic microsatellites in a mangrove species, Rhizophora mangle L. (Rhizophoraceae). Mol Ecol Notes 2:281–283. https://doi.org/10.1046/j.1471-8286.2002.00232.x
Sandoval-Castro E, Dodd RS, Riosmena-Rodriǵuez R et al (2014) Post-glacial expansion and population genetic divergence of mangrove species Avicennia germinans (L.) Stearn and Rhizophora mangle L. along the Mexican coast. PLoS ONE 9:e93358. https://doi.org/10.1371/journal.pone.0093358
Sandoval-Castro E, Muñiz-Salazar R, Enríquez-Paredes LM et al (2012) Genetic population structure of red mangrove (Rhizophora mangle L.) along the northwestern coast of Mexico. Aquat Bot 99:20–26. https://doi.org/10.1016/j.aquabot.2012.01.002
Schwartz MK, Luikart G, Waples RS (2007) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22:25–33. https://doi.org/10.1016/j.tree.2006.08.009
Shafer ABA, Wolf JBW, Alves PC et al (2015) Genomics and the challenging translation into conservation practice. Trends Ecol Evol 30:78–87. https://doi.org/10.1016/j.tree.2014.11.009
Takayama K, Tamura M, Tateishi Y et al (2013) Strong genetic structure over the American continents and transoceanic dispersal in the mangrove genus Rhizophora (Rhizophoraceae) revealed by broad- scale nuclear and chloroplast DNA analysis. Am J Bot 100:1191–1201. https://doi.org/10.3732/ajb.1200567
Takayama K, Tamura M, Tateishi Y, Kajita T (2008) Isolation and characterization of microsatellite loci in the red mangrove Rhizophora mangle (Rhizophoraceae) and its related species. Conserv Genet 9:1323–1325. https://doi.org/10.1007/s10592-007-9475-z
Wee AKS, Mori GM, Lira CF et al (2019) The integration and application of genomic information in mangrove conservation. Conserv Biol 33:206–209. https://doi.org/10.1111/cobi.13140
Wickham H (2011) ggplot2. Wiley Interdiscip Rev Comput Stat 3:180–185. https://doi.org/10.1002/wics.147
Acknowledgements
Permission to collect samples was granted by the Florida Department of Environmental Protection (Permit Number: 07101720), the Department of Marine Resources (The Bahamas), and the Lower Rio Grande Valley National Wildlife Refuge. Many thanks to G Deane, D Devlin, L Duckett, IC Feller, R Feller, L Jupp, L Knowles, and L Simpson for help with sample collections, to S Griffiths for primer development help, to G Fox for providing R-script, and to the University of Manchester Genomic Technologies Core Facility and F Combe for fragment analysis. This work was supported by a Natural Environment Research Council (NERC) EAO Doctoral Training Partnership (NE/L002469/1) to H.C., and The Bahamas National Trust and Bahamian government provided support to A.L. As always, thank you to Clemente and Milo for unconditional support and big smiles.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kennedy, J.P., Craig, H., Jara-Cavieres, A. et al. Multiplex microsatellite PCR panels for the neotropical red mangrove, Rhizophora mangle: combining efforts towards a cost-effective and modifiable tool to better inform conservation and management. Conservation Genet Resour 12, 503–513 (2020). https://doi.org/10.1007/s12686-020-01138-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-020-01138-8