Abstract
Background
Cervical cancer is a prevalent malignancy of the female reproductive system. Cervical intraepithelial neoplasia (CIN) is a precursor lesion for CC. Various studies have examined circulating microRNAs (miRNAs) as potential early diagnostic markers for CC and CIN. However, the findings have been inconclusive. Therefore, it is necessary to evaluate the diagnostic accuracy and identify potential sources of variability among these studies. Methods: The PubMed, Cochrane Library, Embase, and Web of Science databases were searched to identify relevant literature. Then, Stata 14.0 was utilized to calculate summary estimates for diagnostic parameters, including sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the summary receiver operating characteristic (ROC). To scrutinize the heterogeneity, the Cochran-Q test and I2 statistic were utilized. As significant heterogeneity was observed, the random effects model was chosen. To explore potential sources of the heterogeneity, subgroup and regression analyses were conducted. Results: We analysed 12 articles reporting on 24 studies involving 1817 patients and 1731 healthy controls. The pooled sensitivity was 0.77 (95% CI 0.73–0.81), the specificity was 0.81 (95% CI 0.73–0.86), the PLR was 3.99 (95% CI 2.81–5.65), the NLR was 0.28 (95% CI 0.23–0.35), the DOR was 14.18 (95% CI 8.47–23.73), and the area under the curve (AUC) was 0.85 (95% CI 0.81–0.87). Subgroup analysis revealed that multiple miRNAs can improve diagnostic performance; the pooled sensitivity of multiple miRNAs was 0.78 (95% CI 0.68–0.86), the specificity was 0.85 (95% CI 0.78–0.90), and the AUC was 0.89 (95% CI 0.86–0.91). Conclusion: This study suggested that circulating microRNAs may be biomarkers for early CC diagnosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cervical cancer ranked fourth among the leading causes of cancer-related deaths in women worldwide in 2020, with approximately 604,127 new cases and 341,831 deaths [1].
Cervical intraepithelial neoplasia (CIN) is an intermediate stage preceding the onset of cervical cancer. Timely detection and treatment of CIN or early-stage cervical cancer yield more favourable clinical outcomes than treatment of advanced-stage patients. However, with a deficiency in early diagnostic biomarkers, the majority of cervical cancer (CC) cases are identified at later stages [2]. Therefore, prompt recognition of CIN and early-stage CC remains paramount. In addition, current cervical examination techniques, such as HPV DNA testing [3], Papanicolaou (Pap) smear [4], liquid-based cytology (LBC) [5], and colposcopy, are invasive. Hence, there is an immediate need for noninvasive biomarkers to detect CIN and CC.
MicroRNAs (miRNAs), which are approximately 22 nucleotides long, are intrinsic noncoding regulatory RNAs. miRNAs play a pivotal role in modulating physiological and pathological mechanisms by inhibiting or degrading target genes [6]. Recent research has revealed that circulating miRNAs can act as potent biomarkers for various types of cancer, including breast cancer [7], pancreatic cancer [8], non-small cell lung cancer [9], and cervical cancer [10].
Despite an increasing amount of research on the use of circulating miRNAs as diagnostic markers for CIN and CC, recent findings have been inconsistent, and a clinically viable panel for CC diagnosis is lacking. To address these shortcomings, we performed a comprehensive meta-analysis to evaluate the potential diagnostic significance of miRNAs in CIN and CC patients, comparing the diagnostic value of individual miRNAs versus multiple miRNAs.
2 Materials and method
The methodology for this meta-analysis has been registered in the INPLASY.COM, an international database for prospectively registered systematic reviews (INPLASY202340053). The current meta-analysis follows the PRISMA-DTA statement of preferred reporting items for systematic reviews and meta-analyses of diagnostic test accuracy [11].
2.1 Search strategy
Publications were searched in four databases, including PubMed, Embase, Cochrane Library and Web of Science, without language estrictions until November 6, 2022. Utilizing the PICOS framework, our strategic sweep incorporated pertinent Medical Subject Headings (MeSH) and keywords procured from the National Center for Biotechnology Information (NCBI) platform. The detailed search strategy and keywords used are available in Supplementary Material: 1. Moreover, a manual search of relevant articles was also conducted to ensure the thoroughness of the search process.
2.2 Eligibility criteria
The selection criteria mandated: (1) studies pertinent to the diagnostic effectiveness of circulating miRNAs in discerning CIN or CC; (2) inclusion of patients in the case group, diagnosed as per clinically approved criteria; and (3) obtainability of false positive (FP), true positive (TP), false negative (FN), and true negative (TN) frequencies, directly or indirectly. Conversely, any research falling under the exclusionary categories was excluded: (1) cellular, animal, or microbiological experiments; (2) non-comparative studies; and (3) reviews, meta-analyses, or conference summaries.
2.3 Data extraction and quality assessment
Two independent reviewers extracted vital data from articles, covering author details, publication year, ethnicity, miRNAs profiles and expression levels, references, comparison type, sample size of all groups, detection methods of miRNAs, specimen type, AUC with 95% confidence intervals (CIs), and diagnostic performance data (sensitivity, specificity, TP, FP, TN, FN).
Bias risks were appraised utilizing the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool [12]. This instrument encompasses four key areas: patient selection, index test, reference standard, and flow and timing. A third reviewer was involved in when the consensus was not be reached.
2.4 Statistical analysis
For calculating TP, FP, FN, and TN values, we obtained sample size, sensitivity, and specificity from each individual study. Statistical analysis was performed using Stata 14.0 software, including pooled sensitivity, specificity, positive and negative likelihood ratios, diagnostic ratios, and their respective 95% confidence intervals (CIs). Summary receiver operating characteristic (SROC) curves were also plotted to assess the area under the curve. Meta-DiSc 1.4 software was used to examine the threshold effect by evaluating Spearman's correlation coefficient and P value [13]. We appraised the heterogeneity among studies using the Cochran-Q test and I2 statistic for quantitative evaluation. Significant heterogeneity was inferred from a P-value below 0.05 for the Cochran-Q test or an I2 value exceeding 50%, prompting the application of a random-effect model in our analysis. Subgroup analyses and regression analysis served in identifying primary causes of heterogeneity. A sensitivity analysis was conducted to ascertain the reliability and robustness of the meta-analysis outcomes. We utilized Deeks’ funnel plots to check for potential publication bias. Fagan’s nomogram was devised for a more thorough assessment of miRNAs’ diagnostic effectiveness. Review Manager 5.4 assisted in evaluating the literature's quality, and a P-value less than 0.05 was adopted to substantiate the outcomes.
3 Results
3.1 Literature search and study characteristics
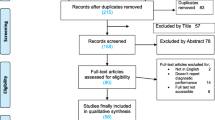
The search of four databases yielded an initial pool of 394 publications. After removing 125 duplicates and excluding 227 irrelevant papers based on the title, study type, and keywords, 42 potential articles remained. Subsequent assessment of the abstracts resulted in further exclusion of 17 papers, leaving 25 full-text papers for further review. Finally, 13 articles were excluded; therefore, 12 eligible articles published between 2014 and 2021 were included in this meta-analysis (Fig. 1 and Table 1).
The 12 included papers comprised 24 case‒control studies. The analysis involved 624 patients with CIN, 1193 patients with CC, and 1731 healthy controls. All of the included studies were published in English, and their primary findings are summarized in Table 2.
Of the 24 studies, 19 focused on a single miRNA, while 5 focused on multiple miRNAs. Serum was the preferred medium for detecting 20 different miRNAs in 38 studies; 3 studies used plasma; and one study used plasma exosomes. Furthermore, miRNA-205 and miRNA-9 were examined in three and two studies, respectively.
3.2 Quality assessment
The QUADAS-2 was used to assess the quality of the 24 studies. As delineated in Fig. 2, every patient was diagnosed with a reference standard, resulting in minimal bias risk for patient diagnosis, as well as patient flow and timing. However, all studies were case‒control studies, leading to an increased risk of selection bias. Moreover, due to their retrospective nature, these studies collectively exhibit a considerable bias risk in the index test.
3.3 Diagnostic accuracy of circulating miRNAs
We chose the random effects model to examine the diagnostic performance of miRNAs for I2 > 50% (I2 = 75.96% for sensitivity and I2 = 89.29% for specificity, Fig. 3). The pooled diagnostic parameters for CIN and CC were as follows: the sensitivity was 0.77 (95% CI 0.73–0.81), the specificity was 0.81 (95% CI 0.73–0.86), the NLR was 0.28 (95% CI 0.23–0.35), the PLR was 3.99 (95% CI 2.81–5.65), and the DOR was 14.18 (95% CI 8.47–23.73). The AUC was 0.85 (95% CI 0.81–0.87) (Fig. 4). There was no evidence of a threshold effect (r = −0.093, P > 0.05).
The clinical applicability of the miRNAs was further evaluated by calculating both the PLR and NLR. High diagnostic accuracy was indicated by a PLR > 10 and NLR < 0.1. Notably, miR-9 and miR-192 showed potential as promising miRNAs for further investigation (Fig. 5A), and a study by Farzanehpour, M., et al. [14] Furthermore, Fagan’s nomogram corroborated the diagnostic accuracy of miRNAs, highlighting posttest probabilities of 50% and 7% for the PLR and NLR, respectively, when the pretest probability was set at 20% (Fig. 5B).
3.4 Subgroup analyses and meta-regression of the diagnostic performance of miRNAs
To further investigate the heterogeneity, we conducted both subgroup (Table 3) and regression analyses (Fig. 6). Subgroup analyses revealed that multiple miRNA profiling and the U6 reference had a significant impact on diagnostic performance. The single miRNAs exhibited sensitivity, specificity, PLR, NLR, DOR, and AUC values of 0.77 (95% CI 0.71–0.82), 0.79 (95% CI 0.70–0.87), 3.73 (95% CI 2.43–5.73), 0.29 (95% CI 0.22–0.38), 12.82 (95% CI 6.80–24.17), and 0.83 (95% CI 0.80–0.86), respectively. Conversely, miRNA panels demonstrated sensitivity, specificity, PLR, NLR, DOR, and AUC values of 0.78 (95% CI 0.68–0.86), 0.85 (95% CI 0.78–0.90), 5.10 (95% CI 3.42–7.61), 0.26 (95% CI 0.17–0.39), 19.76 (95% CI 10.13–38.54), and 0.89 (95% CI 0.86–0.91), respectively.
Moreover, among the selected studies, those using U6 as an internal reference showed a significant diagnostic value compared to the non-U6 group. The U6 subgroup had a sensitivity of 0.82 (95% CI 0.70–0.89), a specificity of 0.87 (95% CI 0.74–0.94), a PLR of 6.14 (95% CI 2.84–13.27), an NLR of 0.21 (95% CI 0.12–0.37), a DOR of 29.17 (95% CI 8.76–97.08), and an AUC of 0.90 (95% CI 0.87–0.93).
We also conducted meta-regression analyses, and the results showed that multiple miRNA profiles, using cervical cancer as the case group, a sample size less than 100, upregulated miRNAs and the use of U6 as a reference affected the sensitivity. Furthermore, using cervical cancer as the case group, miRNAs upregulation and using U6 as the reference affected the specificity.
3.5 Sensitivity analyses
The results of the sensitivity analysis are illustrated in Fig. 7, which shows that the appropriateness of the random effects model was validated based on the goodness of fit (Fig. 7A) and bivariate normality (Fig. 7B). The analysis of influence revealed that the studies by Farzanehpour, M. et al., and Ruan, F. et al. [15] exerted the greatest impact on the topic of weight (Fig. 7C). The identification of outliers in the data from the studies by Farzanehpour,M. et al., and Ruan,F. et al. (Fig. 7D) suggested that the observed heterogeneity may be attributed to these specific data points. Excluding three outlier groups reduced the degree of heterogeneity, as measured by the I2 value, by 3.93% for sensitivity and 3.72% for specificity. Despite excluding outlier groups, no statistically significant change was observed in the degree of heterogeneity (Table 4).
3.6 Publication bias
A funnel plot developed by Deek was employed to visualize the 24 studies included in the analysis. The generated plot, depicted in Fig. 8, exhibited a P-value of 0.87, suggesting no evidence of publication bias.
4 Discussion
CC is one of the predominant gynaecological malignancies. Early detection of CIN and CC is an effective way to reduce the disease burden of cervical cancer. Recently, research has focused on circulating miRNAs as promising biomarkers for the early detection of CIN and CC [16, 17]. Our study aimed to evaluate the diagnostic performance of existing circulating miRNA-based screening methods discussed in the scholarly literature.
Our meta-analysis included 12 papers with 1817 CIN or CC patients and 1731 healthy controls. The results displayed a pooled sensitivity of 0.77 (95% CI 0.73–0.81) and specificity of 0.81 (95% CI 0.73–0.86). A PLR of 3.99 indicates that aberrant miRNA expression substantially increases the accuracy of detection of individuals with CIN and CC, similar to the pooled NLR. This analysis demonstrated a pooled DOR for miRNAs of 124.60, indicating their efficacy in differentiating patients (CC or CIN) from healthy individuals.
In our study, miR-9 and miR-192 showed potential as promising miRNAs for diagnosing CIN and CC. MiR-9 and miR-192 were significantly upregulated in the serum of cervical precancer patients [18]. ROC analysis revealed that miR-9 and miR-192 were highly accurate in discriminating CIN patients from healthy controls, with AUC values of 0.9 and 0.75, respectively [14].
Persistent infection with high-risk human papillomavirus (HPV) strains, particularly HPV-16, HPV-18, HPV-31, HPV-33, HPV-45, HPV-52, and HPV-58, significantly increases the risk of developing cervical lesions and their recurrence after treatment [19, 20]. Bogani's study found that patients with recurrent cervical lesions had longer HPV persistence than those without recurrence (12.2 vs. 9.0 months; p < 0.001) [21]. Additionally, the risk of lesion recurrence after primary conization is 7.46% at 6 months and rises to 13.1% at 12 months if HPV is still detected post-surgery. Moreover, adolescent HPV vaccination can decrease the prevalence of precancerous lesions and cervical cancer. HPV vaccination has been proved to prevent about 70% of lower genital tract dysplasia (LGTD) and reduce the risk of developing new infections of HPV. [22]. Bogani's study showed that vaccinations could offer up to a 67% protective effect [23].
The transformation of normal cervical cells infected with HPV into precancerous cells involves two main processes: the DNA damage response (DDR) and genome amplification of HPV. MiR-9 binds to the 3-UTR of FOXO3 and downregulates FOXO3 expression. FOXO3 inhibits DDR and induces p53-dependent apoptosis. Additionally, miR-9 could be associated with differentiation during the early transformation process. Epithelial-mesenchymal transition (EMT) converts epithelial cells into invasive and migratory mesenchymal cells [24]. The overexpression of miR-9 can lead to EMT, which is regulated by c-Myc and Prospero homeobox 1 (PROX1) [25, 26], contributing to cancer cell metastasis. MiR-192 promotes cell proliferation and migration and decreases apoptosis and cell cycle progression from G0/G1 to S phase by regulating important factors in this process [27, 28]. In colon cancer, it suppresses ZEB2 and VEGFA expression [29]. In renal and ovarian tumour models, it targets ZEB2 and RhoA and mediates TGF-beta-induced EMT [30]. However, its specific targets in cervical cancer remain unknown.
Owing to the heterogeneity of the included studies, we conducted subgroup and meta-regression analyses to examine covariates. Subgroup analysis identified miRNA profiling, comparison type, sample size, miRNA expression, and reference as potential sources of heterogeneity. The multiple miRNAs (AUC value of 0.89) showed better diagnostic accuracy than the single miRNAs (AUC value of 0.84). Our findings align with prior research by Du, SY. et al. [17]
In addition, our findings mandate the selection of suitable internal reference genes for standardization, preferring U6 over cel-39. qRT‒PCR is a common method for profiling circulating miRNA expression, and accurate interpretation depends heavily on selecting appropriate reference genes for normalization. However, the selection of appropriate internal reference genes is still debated [31]. Therefore, identifying consistent and highly stable internal reference genes is important for minimizing bias among tests.
Compared with previous studies [32], we performed subgroup analyses on miRNA regulation modes to investigate the origin of heterogeneity. We found that compared with downregulated miRNAs, upregulated miRNAs exhibited superior diagnostic performance. According to our meta-regression analysis, the regulation mode of miRNAs affects their sensitivity and specificity. Additionally, a similar article by Jiang et al. was published 3 years ago, whereas our study incorporates more recent studies exploring the use of miRNAs in the diagnosis of CC and CIN.
Nevertheless, this analysis has limitations. First, the lack of specified cut-off values for miRNAs in the included studies prevented us from performing a subgroup analysis, which might have resulted in heterogeneity. Second, as all studies included Asian populations, we could not investigate the impact of ethnicity. Third, we included only English language articles. Fourth, the predominance of retrospective case‒control studies increases the risk of selection bias. Last, due to the limited number of similar miRNAs available, it is currently not possible to identify a specific single miRNA or panel of miRNAs as the optimal diagnostic biomarkers for CIN and CC.
Data availability
Not applicable.
References
Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Qiu H, Liang D, Liu L, et al. A Novel circulating MiRNA-based signature for the diagnosis and prognosis prediction of early-stage cervical cancer. Technol Cancer Res Treat. 2020. https://doi.org/10.1177/1533033820970667.
Pontus N, Ryd W, et al. Human papillomavirus and papanicolaou tests to screen for cervical cancer. New Engl J Med. 2007. https://doi.org/10.1056/NEJMoa073204.
Nayar R, Wilbur DC. The bethesda system for reporting cervical cytology: a historical perspective. J Acta Cytologica. 2017;61:359–63.
Whitlock E, Vesco K, Eder M, et al. Liquid-based cytology and human papillomavirus testing to screen for cervical cancer: a systematic review for the US Preventive Services Task Force. Ann Intern Med. 2011. https://doi.org/10.7326/0003-4819-155-10-201111150-00376.
Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–87.
Sehovic E, Urru S, Chiorino G, et al. Meta-analysis of diagnostic cell-free circulating microRNAs for breast cancer detection. BMC Cancer. 2022;22:634.
Ebrahimi S, Hosseini M, Ghasemi F, et al. Circulating microRNAs as potential diagnostic, prognostic and therapeutic targets in pancreatic cancer. Curr Pharm Des. 2016;22:6444–50.
Hetta HF, Zahran AM, Shafik EA, et al. Circulating miRNA-21 and miRNA-23a expression signature as potential biomarkers for early detection of non-small-cell lung cancer. MicroRNA (Shariqah, United Arab Emirates). 2019;8:206–15.
Lv A, Tu Z, Huang Y, et al. Circulating exosomal miR-125a-5p as a novel biomarker for cervical cancer. Oncol Lett. 2021;21:54.
Mcgrath TA, Alabousi M, Skidmore B, et al. Recommendations for reporting of systematic reviews and meta-analyses of diagnostic test accuracy: a systematic review. systematic review. 2017;6:194.
Whiting PF, Rutjes A, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36.
Zamora J, Abraira V, Muriel A, et al. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Resea Methodol. 2006;6:1–12.
Farzanehpour M, Mozhgani S-H, Jalilvand S, et al. Serum and tissue miRNAs: potential biomarkers for the diagnosis of cervical cancer. Virol J. 2019. https://doi.org/10.1186/s12985-019-1220-y.
Ruan F, Wang YF, Chai Y. Diagnostic values of miR-21, miR-124, and M-CSF in patients with early cervical cancer. Technol Cancer Res Treat. 2020. https://doi.org/10.1177/1533033820914983.
Ning R, Meng S, Wang L, et al. 6 circulating miRNAs can be used as non-invasive biomarkers for the detection of cervical lesions. J Cancer. 2021;12:5106–13.
Du S, Zhao Y, Lv C, et al. Applying serum proteins and MicroRNA as novel biomarkers for early-stage cervical cancer detection. Sci Rep. 2020;10:9033.
Xin F, Liu P, Ma CF. A circulating serum miRNA panel as early detection biomarkers of cervical intraepithelial neoplasia. Eur Rev Med Pharmacol Sci. 2016;20:4846–51.
Bogani G, Sopracordevole F, Ciavattini A, et al. HPV persistence after cervical surgical excision of high-grade cervical lesions. Cancer Cytopathol. 2023. https://doi.org/10.1002/cncy.22760.
Lax S. Histopathology of cervical precursor lesions and cancer. Acta Dermatovenerol Alp Pannonica Adriat. 2011;20:125–33.
Bogani G, Sopracordevole F, Ciavattini A, et al. Duration of human papillomavirus persistence and its relationship with recurrent cervical dysplasia. Eur J Cancer Prev. 2023;32(6):525–32.
Waheed DEN, Burdier FR, Eklund C, et al. An update on one-dose HPV vaccine studies, immunobridging and humoral immune responses—a meeting report. Prev Med Rep. 2023;35: 102368.
Bogani G, Sopracordevole F, Ciavattini A, et al. HPV-related lesions after hysterectomy for high-grade cervical intraepithelial neoplasia and early-stage cervical cancer: a focus on the potential role of vaccination. Tumori. 2023;110(2):139–45.
Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54.
Lu MH, Huang CC, Pan MR, et al. Prospero homeobox 1 promotes epithelial-mesenchymal transition in colon cancer cells by inhibiting E-cadherin via miR-9. Clin Cancer Res. 2012;18:6416–25.
Khew-Goodall Y, Goodall GJ. Myc-modulated miR-9 makes more metastases. Nat Cell Biol. 2010;12:209–11.
Galina FG, Avital G, Keren S, et al. miR-192 directly binds and regulates dicer1 expression in neuroblastoma. PLoS ONE. 2013;8: e78713.
Zhao C, Zhang J, Zhang S, et al. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol Rep. 2013;30:276–84.
Geng L, Chaudhuri A, Talmon G, et al. MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene. 2014;33:5332–40.
Wu SY, Rupaimoole R, Shen F, et al. A miR-192-EGR1-HOXB9 regulatory network controls the angiogenic switch in cancer. Nat Commun. 2016;7:11169.
Donati S, Ciuffi S, Brandi ML. Human circulating miRNAs Real-time qRT-PCR-based analysis: an overview of endogenous reference genes used for data normalization. Int J Mol Sci. 2019. https://doi.org/10.3390/ijms20184353.
Jiang Y, Hu ZH, Zuo ZH, et al. Identification of circulating MicroRNAs as a promising diagnostic biomarker for cervical intraepithelial neoplasia and early cancer: a meta-analysis. BioMed Res Int. 2020. https://doi.org/10.1155/2020/4947381.
Cao S, Li H, Li L. LncRNA SNHG17 contributes to the progression of cervical cancer by targeting microRNA-375-3p. Cancer Manag Res. 2021;13:4969–78.
Juan L, Tong HL, Zhang P, et al. Identification and characterization of novel serum microRNA candidates from deep sequencing in cervical cancer patients. Sci Rep. 2014;4:6277.
Kong Q, Tang Z, Xiang F, et al. Diagnostic value of serum hsa-MIR-92a in patients with cervical cancer. Clin Lab. 2017;63:335–40.
You W, Wang Y, Zheng J. Plasma MIR-127 and MIR-218 might serve as potential biomarkers for cervical cancer. Reprod Sci. 2015;22:1037–41.
Zhang YJ, Zhang DH, Wang F, et al. Serum miRNAs panel (miR-16–2*, miR-195, miR-2861, miR-497) as novel non-invasive biomarkers for detection of cervical cancer. Sci Reports. 2015. https://doi.org/10.1038/srep17942.
Zheng S, Li R, Liang J, et al. Serum miR-638 combined with squamous cell carcinoma-related antigen as potential screening biomarkers for cervical squamous cell carcinoma. Genet Test Mol Biomarkers. 2020;24:188–94.
Funding
This work was supported by National Natural Science Foundation of China (81702686, 81602920 and 82102850), National Natural Science Foundation of Jiangsu (BK20211383), Nanjing medical science and technology development foundation (ZKX21048). Cancer Precision Radiotherapy Spark Program of China International Medical Foundation, grant number (2019-N-11-12). Jiangsu cancer hospital funding (ZM202007).
Author information
Authors and Affiliations
Contributions
Yue Li and Longbiao Zhu conducted the investigation by processing data and writing, Chenjing Zhu, Hangju Zhu and Yan Chen checked data, Hui Yu, Ping Yin and Mengyu Liu checked the figures, Yang Li and Huixin Li checked the figures, Jing Han was the project administration and supervised the project, Hanzi Xu proposed the project and funded the project, Zhen Gong edited the writing. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Consent to participate
All participants signed written informed consent.
Competing interests
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Y., Zhu, L., Zhu, C. et al. Circulating micrornas as potential diagnostic biomarkers for cervical intraepithelial neoplasia and cervical cancer: a systematic review and meta-analysis. Discov Onc 15, 189 (2024). https://doi.org/10.1007/s12672-024-01028-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-024-01028-7