Abstract
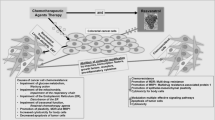
Cancer resistance to chemotherapeutic agents is a major issue in the management of cancer patients. Overexpression of the ribonucleotide reductase regulatory subunit M2 (RRM2) has been associated with aggressive cancer behavior and chemoresistance. Nano-diamino-tetrac (NDAT) is a nanoparticulate derivative of tetraiodothyroacetic acid (tetrac), which exerts anticancer properties via several mechanisms and downregulates RRM2 gene expression in cancer cells. Resveratrol is a stilbenoid phytoalexin which binds to a specific site on the cell surface integrin αvβ3 to trigger cancer cell death via nuclear translocation of COX-2. Here we report that resveratrol paradoxically activates RRM2 gene expression and protein translation in colon cancer cells. This unanticipated effect inhibits resveratrol-induced COX-2 nuclear accumulation. RRM2 downregulation, whether achieved by RNA interference or treatment with NDAT, enhanced resveratrol-induced COX-2 gene expression and nuclear uptake which is essential to integrin αvβ3-mediated-resveratrol-induced antiproliferation in cancer cells. Elsewhere, NDAT downregulated resveratrol-induced RRM2 expression in vivo but potentiated the anticancer effect of the stilbene. These findings suggest that RRM2 appears as a cancer cell defense mechanism which can hinder the anticancer effect of the stilbene via the integrin αvβ3 axis. Furthermore, the antagonistic effect of RRM2 against resveratrol is counteracted by the administration of NDAT.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer resistance to chemotherapeutic agents is a major issue in the management of cancer patients. More than their normal counterparts, cancer cells possess a large panel of cell defense pathways that enable adaptive behaviors to new hostile environments induced by drugs. While some cancer cells exert a primary resistance to certain drugs, other tumors express survival strategies that progressively render treatments ineffective. Various mechanisms are utilized, including drug inactivation, drug target alteration, drug export/efflux, DNA damage repair, and anti-apoptosis [1]. Chemoresistance is one rationale for anticancer combination therapy, as different drugs combined can synergistically tackle several mechanisms to disorganize cancer cells’ defense systems.
Ribonucleoside-diphosphate reductase subunit M2—the gene product of ribonucleotide reductase regulatory subunit M2 (RRM2)—is the small subunit of the ribonucleotide reductase (RNR) which catalyzes the rate-limiting synthesis of deoxyribonucleoside triphosphates (dNTPs) required for DNA replication and repair [2]. Overexpression of RRM2 is associated with cancer cell proliferation. Indeed, increased expression of RRM2 has been correlated with aggressive behavior in bladder cancer [3], invasion of muscularis propria and higher Ki-67 labeling index in gastric cancers [4], proliferation and invasiveness in breast cancer [5], and poor survival in colorectal cancers [6]. Moreover, the DNA-repair function of RRM2 is highjacked by cancer cells to activate resistance against chemotherapeutic agents. For instance, breast cancer cells leverage RRM2 to induce tamoxifen resistance via overexpression of AKT [7]. Elsewhere, RRM2 overexpression mediates pancreas adenocarcinoma cells’ resistance to gemcitabine [8, 9].
Integrin αvβ3 is a proneoplastic integrin amply expressed in cancer cells and rapidly dividing endothelial cells [10]. The extracellular domain of this integrin provides binding sites for several hormone-like molecules including resveratrol, dihydrotestosterone, and thyroid hormones and their derivatives, all of which dictate different behaviors in cancer cells [11, 12]. For instance, the thyroid hormone L-thyroxine (T4), via binding on αvβ3, initiates non-genomic mechanisms which culminate in cancer cell nucleus-mediated events, such as cell proliferation, survival, angiogenesis, and metastasis [13, 14]. In contrast, tetraiodothyroacetic acid (tetrac) is a deaminated analog of T4, which, along with its nanoparticulate analog, Nanotetrac (nano-diamino-tetrac, NDAT), impairs cancer cell proliferation both in vitro and in xenograft animal models [10, 15, 16]. On integrin αvβ3, NDAT and tetrac competitively inhibit the binding of T4, blocking cancer cell signals initiated by this thyroid hormone. However, independently of binding competition with T4, NDAT and tetrac affect cancer cell functions and exert anticancer properties via several mechanisms. These include inhibition of cancer cell proliferation, anti-angiogenesis, suppression of metastasis, promotion of cancer cell apoptosis [16,17,18], and inhibition of immune system-escape mechanisms (immune checkpoints) of cancer cells [15]. Importantly, some thyromimetic (i.e., genomic) effects have been observed with tetrac which is internalized by cancer cells [19]. Hence, its nanoparticulate analog has been engineered; it is composed of tetrac molecules linked to a PLGA nanoparticle to prevent cellular uptake of any modified tetrac that gains access to the cell interior [20]. Notably, T4 upregulates RRM2 expression in human colon cancer cells, while NDAT prevents RRM2 gene expression and also impairs T4-induced RRM2 expression [21].
Resveratrol, a stilbenoid polyphenolic antioxidant, has been shown to induce apoptosis in different cancer cell lines and to suppress cancer growth in animal models [16]. We have shown that resveratrol (1–10 μM), acting at its receptor on integrin αvβ3, induces p53-dependent apoptosis in different cancer cells [22, 23]. Resveratrol activates the extracellular signal-regulated kinase 1 and 2 (ERK1/2) pathway and triggers the nuclear accumulation of an inducible form of COX-2. Nuclear COX-2 and phosphorylated ERK1/2 cooperate to activate p53 via phosphorylation of serine 15 and subsequent antiproliferation in different cancer cells [22].
In this study, we thought to investigate the effect of resveratrol on RRM2 gene expression and protein translation, as well as ascertain the potentiating effect of NDAT on the stilbene’s anticancer function. If NDAT does so, we wanted to explore the mechanisms involved in vitro and in an animal xenograft model.
Materials and Methods
Materials
NDAT was synthesized as previously described [20]. Thyroid hormone and resveratrol were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA). Resveratrol was prepared as a stock concentration of 100 nM in dimethyl sulfoxide (DMSO). NDAT and T4 were prepared as stock concentrations of 10−2 M in PBS and KOH-propylene glycol, respectively. Reagents were conserved at − 20 °C. The final concentrations of solvents used to dissolve the reagents were tested for activity and did not affect the results of the experiments.
Cell Cultures
HCT 116 (ATCC) and HT-29 (ATCC) human colorectal cancer cell lines were acquired from the Bioresource Collection and Research Center (BCRC, Hsinchu, Taiwan) which purchased them from the American Type Culture Collection (ATCC) and proceeded with their authentication. The cells were cultured in RPMI 1640 medium containing 10% FBS, penicillin 100 U/ml, and streptomycin 100 μg/ml, under 5% CO2/95% air at 37 °C. The cells were used for experiments until passage 15. Prior to treatment, the cells were placed in serum-free medium for starvation during 48 h. The cells were treated with drugs or hormones in 1% hormone-stripped FBS-containing medium.
Immunoblotting
Total proteins were extracted by lysing cells in radioimmunoprecipitation assay (RIPA) buffer containing onefold protease inhibitor and onefold phosphatase inhibitor. Protein nuclear fractions were extracted according to the Thermo Scientific NE-PER® Nuclear and Cytoplasmic Extraction Kit protocol (Thermo Scientifc). The blotting techniques have been described in several of our previous papers [15, 22]. Primary antibodies used were as follows: COX-2 (Cayman aa 584-598 1:2000), RRM2 (Abcam ab57653, 1–5 μg/ml), p53(GeneTex, GTX102965, 1:1000), pSer15-p53 (GeneTex, GTX21431, 4 μg/ml), ERK2 (GeneTex, GTX113094, 1:1000), pERK1/2 (Cell signaling ref. 06|2016 43775, 1:1000), GAPDH (GeneTex GTX100118, 1:1000), and Lamin B1 (GeneTex, GTX103292, 1:1000). Depending upon the origin of the primary antibody, the secondary antibodies were rabbit anti-mouse IgG, goat anti-rabbit IgG, or rabbit anti-goat horseradish peroxidase diluted at 1:2000. Amersham™ Imager 600 imaging systems (GE Healthcare) was used to detect immunoreactive proteins. Blots were analyzed using ImageJ 1.5 software (NIH, USA).
Real-Time PCR
After customized treatment, total RNA was extracted following the illustra RNAspin Mini RNA Isolation Kit (GE Healthcare Life Sciences) protocol. Two micrograms of RNA was treated with DNase I and used as templates for reverse transcription into cDNA, with RevertAid H Minus First Strand cDNA Synthesis Kit (Life Technologies). The resulting cDNA was used to perform real-time PCR reactions on CFX Connect™ Real-Time PCR Detection System (Bio-Rad Laboratories) according to the QuantiNova™ SYBR® Green PCR Kit (QIAGEN) protocol described in our previous studies [22]. The gene primers and accession numbers are shown in Table 1.
Cell Proliferation Assay
Cells (5 × 103 cells per well) were seeded on 6-well trays and subjected to starvation for 48 h in serum-free RPMI 1640 medium. Next, the cells were treated for 72 h in RPMI 1640 containing 1% charcoal-stripped FBS. Medium and agents were renewed on a daily basis. Finally, the cells were harvested after tripsin treatement and counted using Countess™ automated cell (Invitrogen, Thermo Fisher Scientifc).
RNA Interference
Human HCT 116 and HT-29 colon cancer cells were seeded in 6-well trays (105 cells/well). Next, cells were grown to 60~80% confluence in an antibiotic-free medium for 24 h before transfection. Cells were then transfected with RRM2 shRNA (Academia Sinica, Core Facility), using Lipofectamine 3000, in Opti-MEM I medium according to the manufacturer’s instructions. Transfected cells were cultured at 37 °C for 4 h and placed in fresh culture medium for an additional 48 h. Finally, transfected cells were treated or not, with NDAT 10−7 M, resveratrol 10 μM, or their combination, for 12 h (qPCR) or 24 h (Western blot analysis), then harvested for subsequent experiments.
Immunohistochemistry
Formalin-fixed and paraffin-embedded tumor sections were examined for RRM2 expression using Novolink Polymer Detection System (Novocastra Laboratories, Wetzlar, Germany). The complete protocol has been described in our previous studies [16]. Tissue sections were incubated overnight with RRM2 (Abcam ab57653, 5 μg/ml). Images were examined under the Nikon Eclipse Ci optical microscope imaging system (Nikon Eclipse Ci, Nikon Instruments) and captured at ×400 magnification, scale bar of 50 μm.
Confocal Microscopy
Exponentially growing human colon HCT 116 cancer cells were seeded on sterilized cover glasses (Paul Marienfeld, Lauda-Königshofen, Germany) and subjected to starvation for 48 h, prior to being treated with agents for 24 h. Supplementary details are provided in our previous work [16]. Cell were incubated overnight with a monoclonal antibody to RRM2 (Abcam ab57653; 1:500), followed by an incubation with Alexa-647-labeled goat anti-mouse (ab150119; 1:500). The fluorescent signals from RRM2 were recorded and analyzed with the TCS SP5 Confocal Spectral Microscope Imaging System (Leica Microsystems). The figures shown are representative of five fields for each experimental condition.
Animal Studies
Immunocompromised nude mice were purchased from the National Laboratory Animal Center (Taipei, Taiwan) and housed in a reserved, pathogen-free facility, in accordance with the protocols approved by the Institutional Animal Care and Use Committee of National Defense Medical Center, Taipei, Taiwan (IACUC-15-340). Mice were acclimatized to our vivarium for 1 week prior to their use. Two to three animals were housed in a cage under conventional conditions and fed ad libitum.
Xenografts
Mice were inoculated with 2 × 107 human colon cancer HCT 116 cells. After the tumor onset (10 days post-inoculation), mice were randomly allocated to the following groups of five: control, NDAT 0.1 mg/kg, resveratrol 25 mg/kg and NDAT 0.1 mg/kg + resveratrol 25 mg/kg. All agents were administered in the peritoneum. Resveratrol was injected 5 days a week and NDAT, twice a week (Monday and Friday). Resveratrol was diluted in 1% Tween 80 for better solubility. The tolerance to agents was monitored every day while the tumor volume and the animals’ weight were evaluated once a week. On day 21, the animals were sacrificed; a section of tumor tissues was fixed in formalin and then embedded in paraffin for immunohistochemistry. Additional fractions were frozen in liquid nitrogen and stored at − 80 °C for qPCR experiments.
Statistical Analysis
Statistical analysis was conducted with IBM SPSS Statistics software, version 20.0 (SPSS Inc., Chicago, IL, USA). A one-way analysis of variance was performed for multiple comparisons between groups, followed by a Bonferonni post hoc. p value < 0.05 was considered statistically significant.
Results
Thyroid Hormone T4 Blocks Actions of Resveratrol in Colorectal Cancer Cells
Thyroid hormone T4 has been shown to induce ERK1/2 phosphorylation and cancer cell proliferation [24]. To ascertain the action of T4 on resveratrol-induced apoptosis in colon cancer, we treated human colorectal cancer HCT 116 and HT-29 cells with 10 μM of resveratrol, 10−7 M of T4, or a combination of both agents. Resveratrol indeed stimulated ERK1/2 phosphorylation, COX-2 nuclear translocation, and p53 phosphorylation at serine 15 (Fig. 1a). T4 at 10−7 M, alone, caused ERK1/2 phosphorylation but did not affect COX-2 nuclear accumulation or p53-Ser15 phosphorylation (Fig. 1a). Furthermore, resveratrol-induced COX-2 nuclear accumulation and p53-Ser15 phosphorylation were blocked by T4 (Fig. 1a). At the gene level, resveratrol promoted CDKN1A (p21), TP53, and COX-2 gene expression, which was hindered upon T4 co-treatment (Fig. 1b). Likewise, T4 stimulated colon cancer cell proliferation and significantly compromised the anticancer effect of resveratrol in the combination regimen (Fig. 1c). Taken together, these results indicated that in colon cancer cells, T4 inhibits resveratrol’s anticancer functions, an effect which depends in part upon the prevention of resveratrol-induced COX-2 nuclear accumulation.
Thyroid hormone T4 blocks actions of resveratrol in colorectal cancers. Following 48 h of starvation, human colorectal cancer HCT 116 and HT-29 cells were treated with 10 μM resveratrol, 10−7 M of T4, or the combination of both agents in serum containing 1% stripped FBS. a Nuclear proteins were extracted after 24 h of treatment for Western blot analyses and normalized to nuclear Lamin B1. Representative blots of three different experiments are presented. b Total RNA was extracted after 24 h of treatment and qPCR experiments performed for COX-2, p21, and TP53 gene expression. c Cells were seeded in 6-well strays and treated for 72 h, then harvested and counted. Agents and medium were renewed on a daily basis. Data are displayed as the mean ± SD of at least three experiments run as triplicates. *,#p < 0.05; **,##p < 0.01; ***,###p < 0.001
NDAT Promotes Resveratrol-Induced Apoptosis in Colorectal Cancer Cells
NDAT prevents T4 from binding to integrin αvβ3 and blocks the signal transduction pathways initiated by the hormone at this plasma membrane receptor [13, 25]. In order to examine the potentiating effect of NDAT on resveratrol-induced antiproliferation in colorectal cancer cells, KRAS mutant but APC wild-type HCT 116 cells (Fig. 2a) and KRAS wild-type but APC mutant HT-29 cells (Fig. 2b) were treated with resveratrol (1 or 10 μM), NDAT (10−7 M), alone, or combined with resveratrol for 72 h. Both agents significantly inhibited cancer cell proliferation. However, the effect of the combination treatment was greater than a single agent at the same concentration. More importantly, results were similar regardless of the KRAS and APC mutation status of the cells (Fig. 2a, b).
NDAT potentiates resveratrol-induced antiproliferation in colorectal cancers. Following 48 h of starvation, human colorectal cancer HCT 116 (a) and HT-29 (b) cells were treated with resveratrol (1 or 10 μM), NDAT 10−7 M alone, or in combination with resveratrol, in serum containing 1% stripped FBS. Agents and medium were renewed on a daily basis. Cells were harvested after 72 h and cell numbers were counted. The results of three different experiments run as triplicates are presented. Data are displayed as the mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with control, ###p < 0.001 compared with NDAT alone, and $$$p < 0.001 compared with resveratrol alone at similar concentration
Resveratrol and NDAT Oppositely Modulate RRM2 Expression in Colon Cancer Cells, but NDAT Enhances Resveratrol-Induced COX-2 Gene Expression and Protein Nuclear Translocation
The oncogenic properties of RRM2 have been extensively described [5, 6]. Consequently, we sought to determine the effect of resveratrol on RRM2 gene expression and protein translation in colon cancer cells. For this purpose, human colon cancer HCT 116 cells were exposed to different concentrations of resveratrol for 12 h. Cells were harvested and qPCR experiments were performed. Unexpectedly, resveratrol induced RRM2 gene expression, which was concentration-dependent (Fig. 3a). A similar pattern was observed in HT-29 colon cancer cells (Fig. 3a). In contrast, NDAT downregulated RRM2 gene expression in both cell lines; moreover, NDAT significantly impaired the induction of RRM2 expression by resveratrol (Fig. 3b). However, NDAT promoted resveratrol-induced COX-2 gene expression (Fig. 3c). At the protein level, 24 h exposure to resveratrol also enhanced RRM2 abundance (Fig. 3d). NDAT, in contrast, significantly decreased RRM2 accumulation and blocked resveratrol-induced RRM2 protein translation in the combination treatment (Fig. 3d). Furthermore, resveratrol-induced COX-2 protein nuclear translocation was enhanced by NDAT co-treatment (Fig. 3d).
Effect of resveratrol and NDAT on RRM2 and COX-2 expression in colorectal cancers. a–c Human colorectal cancer HCT 116 and HT-29 cells were treated with agents for 12 h after 48 h of starvation, and gene expression of RRM2 and COX-2 were detected by qPCR experiments. d After 24 h of treatment, total proteins were collected for RRM2 expression normalized to GAPDH and nuclear proteins for COX-2 normalized to Lamin B1. Data are displayed as the mean ± SD of three different experiments. *,#p < 0.05; **,##p < 0.01; ***,###p < 0.001
To further explore the effect of resveratrol and NDAT on RRM2 expressions, we conducted confocal microscopy experiments on HCT 116 cancer cells after 24 h of exposure to agents. Cytoplasmic staining showed that RRM2 amount (green fluorescence) was enhanced by resveratrol but decreased in the presence of NDAT both alone and in the combination treatment (Fig. 4).
Confocal microscopy of RRM2 upon agents treatment. Human colon cancer HCT 116 cells were treated with NDAT 10−7 M, resveratrol 10 μM, or their combination for 24 h, and cells were examined by confocal microscopy. NDAT downregulated RRM2 protein (green fluorescence) concentration and reduced resveratrol-induced RRM2 accumulation in the combination treatment
RRM2 Knockdown Enhances Resveratrol-Induced COX-2 Gene Expression and Protein Nuclear Accumulation
The unexpected finding that resveratrol induces RRM2 expression at both gene and protein levels led us to further examine the involvement of RRM2 in resveratrol’s anticancer functions. To this end, RNA interference experiments were conducted on human colon cancer HCT 116 and HT-29 cells. Results indicated that RRM2 knockdown potentiated resveratrol-induced COX-2 gene expression (Fig. 5a, b). Western blot analyses further confirmed that RRM2 silencing promoted resveratrol-induced COX-2 nuclear translocation (Fig. 5c). These observations indeed suggested that somehow, RRM2 compromises resveratrol’s anticancer abilities in colon cancer by hindering resveratrol-induced COX-2 nuclear accumulation.
RRM2 downregulation promotes resveratrol-induced COX-2 gene expression and protein nuclear translocation. a, b Human colorectal cancer cell lines were transfected with shRNA of RRM2 for 48 h, then were treated with 10 μM resveratrol, NDAT 10−7 M, or their combination for another 12 h for qPCR. Data are displayed as the mean ± SD of three experiments. *p < 0.05, **p < 0.01, ***p < 0.001; ###p < 0.001. c Nuclear proteins were extracted after 24h treatment, for Western blot experiments. COX-2 protein is normalized to Lamin B1. Data are displayed as the mean ± SD of three experiments. **p < 0.01, ***p < 0.001 compared to control; ##p < 0.01, ###p < 0.001 compared to resveratrol 10 μM; $$$p < 0.001 compared to shRRM2 control
NDAT Promotes Resveratrol-Induced Antiproliferation In Vivo
To characterize the effects of NDAT and resveratrol on gene expression in vivo, we performed qPCR experiments on xenograft tumor samples. These experiments revealed that resveratrol increased RRM2 gene expression, an action that is impaired by NDAT co-treatment (Fig. 6a). Additionally, resveratrol-induced CDKN1A (p21), TP53, and COX-2 gene expression was promoted by NDAT co-administration (Fig. 6a).
Effect of NDAT and resveratrol in mouse xenograft model. Human colon cancer HCT 116 cells (2 × 107) were injected into the flanks of nude mice as described in the “Materials and Methods.” a qPCR experiments of tumor samples. b IHC staining: microscopic photographs of representative specimens are shown (control, NDAT 0.1 mg/kg, resveratrol 25 mg/kg, or combination treatment). RRM2 protein is stained in brown color
We next performed immunohistochemistry experiments on tumor samples. RRM2 protein was increased in the resveratrol-treated groups compared to the control group, whereas NDAT—alone or in combination with resveratrol—hindered RRM2 protein translation (Fig. 6b). Altogether, these results suggested that RRM2 interfers with resveratrol’s antitumoral effect in vitro and in vivo, and this action of resveratrol can be rescued with NDAT co-administration.
To determine whether NDAT potentiates the overall anticancer effect of resveratrol in vivo, we studied a nude mice xenograft model of colon cancer. Treatment of xenografted animals with either NDAT or resveratrol significantly inhibited tumor growth (Fig. 7a, b and Table 2). NDAT at 0.1 mg/kg yielded an inhibition rate of 76.98%, comparable to resveratrol at 25 mg/kg (80.99%). More importantly, the potentiating effect of NDAT on resveratrol-induced antiproliferation was reproducible in vivo, as the combination treatment induced a more significant antitumoral effect (Table 2). These agents were well tolerated by animals and caused no loss of body weight (Fig. 7c).
Effect of NDAT and resveratrol on cancer growth in vivo. a Growth curve of tumor volume normalized to the volume at the beginning of the treatment. b Between group comparison of final tumor volumes. c Animal body weight at the beginning and the end of the treatment. Data are shown as the mean ± SD. **p < 0.01 compared to control, $ p<0.05 compared to NDAT 0.1 mg/kg, ##p<0.01 compared to resveratrol 25 mg/kg
Discussion
In this study, we showed that resveratrol paradoxically triggers RRM2 expression, which impedes the anticancer effect of the stilbene via the integrin αvβ3 axis. NDAT abrogated resveratrol-induced RRM2 gene expression and protein translation. Comparatively, RRM2 downregulation achieved in the present studies by RNA interference potentiated resveratrol-induced COX-2 gene expression and protein nuclear translocation, which—in cancer cells—is shown as being fundamental for integrin αvβ3-mediated resveratrol-induced antiproliferation [12, 23]. These results suggested that the induction of RRM2 expression by resveratrol is detrimental to the latter’s anticancer properties. Both resveratrol and NDAT exert an inhibitory effect on cancer cell proliferation in vitro and in vivo [16, 26]. However, the combination of resveratrol and NDAT had an antiproliferation effect that exceeds that of single agents, implying a synergistic effect between the two drugs.
One large subunit (M1) binds to either one of two small subunits (RRM2 and RRM2B) to form the heterodimeric ribonucleoside-diphosphate reductase [27]. RRM2B has been identified as an inducible form of RRM2 which is synthesized in response to wild-type p53 activation [28]. On the other hand, RRM2 overexpression has been consistently associated with cancer aggressive behavior. More importantly, increase in RRM2 function supports cancer defense mechanisms against chemotherapeutic agents [7, 29]. In the current study, we found that RRM2 was upregulated in colon cancer cells in response to certain concentrations of resveratrol. Because resveratrol has well-described anticancer properties, we had expected this stilbene to downregulate RRM2. Therefore, RRM2 response to resveratrol in cancer cells appears to be an adaptive mechanism of these cells to the anticancer activity of the stilbene. Indeed, downregulation of RRM2 restored resveratrol-induced COX-2 gene expression and protein nuclear translocation. These are steps in the pro-apoptotic activity of resveratrol. Upregulation of oncogenic signals such as the Akt pathway [7] determines RRM2 overexpression and the ensuing chemoresistance. The mechanisms involved in this increase of RRM2 under resveratrol treatment have yet to be determined.
Similarly to other naturally derived anticancer agents, resveratrol is a multi-target drug which affects several key cell signaling systems [30]. That resveratrol can unexpectedly promote oncogenic molecules has already been described. Using the human fibrosarcoma cell line HT1080, Gweon and Kim reported that resveratrol induces MMP9 expression which was mediated via p38 kinase and PI-3K pathways [31]. Although the overall cancer cell viability was compromised by resveratrol treatment, this increased expression of MMP9 still supports cancer cell migration and metastasis. Elsewhere, epigallocatechin-3-gallate (EGCG), a green tea polyphenol which has been described to exhibit chemopreventive effects, has also been found to adversely induce pro-MMP7 mRNA and protein and to generate superoxide O2− [32]. These reports and our current work indeed point out that these agents are pleiotropic and can yield undesirable effects. Nonetheless, these observed effects are overtaken by their anticancer abilities.
We have previously shown that tetrac downregulates both high-mobility group AT-hook 2 (HMGA2) and β-catenin in colon cancer, and this phenomenon similarly promoted resveratrol-induced COX-2 nuclear accumulation and the subsequent anticancer effect in colon cancer cells HCT 1116 and HT-29 [16]. In the pathogenesis of colon cancer, β-catenin and KRAS play key roles [33, 34]. Interestingly, there is crosstalk between both of these abovementioned oncogenes and RRM2 in cancer cells. Indeed, KRAS acts upstream to upregulate RRM2 in human colon cancer HCT 116 cells, leading to increased cancer cell proliferation [29]. RRM2 is also known to utilize β-catenin to promote oral cancer invasiveness [35]. In our study, we found that NDAT downregulated RRM2 expression in colon cancer cells regardless of the status of APC and KRAS. NDAT antagonizes on T4 and T4-dependent PD-L1 and RRM2 expressions in colon cancer cells [15, 21]. Both T4 and PD-L1 have been shown to prevent resveratrol-induced antiproliferation in cancer cells [23]. Indeed, T4 prevents resveratrol-induced COX-2 nuclear accumulation that contributes to apoptosis caused by resveratrol [36] (Fig. 1). In addition, we recently found that T4 induces PD-L1 in ovarian cancer cells. PD-L1 in turn trapped COX-2 in the cytoplasm and prevented COX-2 nuclear uptake upon treatment with resveratrol [37]. Therefore, several mechanisms exist in cancer cells by which T4 may oppose resveratrol’s anticancer properties. NDAT shares structural homologies with T4 and competitively inhibits the binding of T4 to its specific receptor on integrin αvβ3 [38]. This new finding that RRM2 is upregulated by resveratrol highlights an additional advantage of employing NDAT in combination therapy, especially in settings where high levels of T4 may promote cancer progression.
References
Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S (2014) Drug resistance in cancer: an overview. Cancers (Basel) 6(3):1769–1792
Nordlund P, Reichard P (2006) Ribonucleotide reductases. Annu Rev Biochem 75:681–706
Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M (2010) Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology 57(6):885–892
Morikawa T, Hino R, Uozaki H, Maeda D, Ushiku T, Shinozaki A, Sakatani T, Fukayama M (2010) Expression of ribonucleotide reductase M2 subunit in gastric cancer and effects of RRM2 inhibition in vitro. Hum Pathol 41(12):1742–1748
Zhang H, Liu X, Warden CD, Huang Y, Loera S, Xue L, Zhang S, Chu P, Zheng S, Yen Y (2014) Prognostic and therapeutic significance of ribonucleotide reductase small subunit M2 in estrogen-negative breast cancers. BMC Cancer 14:664
Liu X, Zhang H, Lai L, Wang X, Loera S, Xue L, He H, Zhang K, Hu S, Huang Y, Nelson RA, Zhou B, Zhou L, Chu P, Zhang S, Zheng S, Yen Y (2013) Ribonucleotide reductase small subunit M2 serves as a prognostic biomarker and predicts poor survival of colorectal cancers. Clin Sci (Lond) 124(9):567–578
Shah KN, Mehta KR, Peterson D, Evangelista M, Livesey JC, Faridi JS (2014) AKT-induced tamoxifen resistance is overturned by RRM2 inhibition. Mol Cancer Res 12(3):394–407
Duxbury MS et al (2003) RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene 23(8):1539–1548
Minami K, Shinsato Y, Yamamoto M, Takahashi H, Zhang S, Nishizawa Y, Tabata S, Ikeda R, Kawahara K, Tsujikawa K, Chijiiwa K, Yamada K, Akiyama SI, Pérez-Torras S, Pastor-Anglada M, Furukawa T, Yasuo T (2015) Ribonucleotide reductase is an effective target to overcome gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells with dual resistant factors. J Pharmacol Sci 127(3):319–325
Davis PJ et al (2014) Cancer cell gene expression modulated from plasma membrane integrin αvβ3 by thyroid hormone and nanoparticulate tetrac. Front Endocrinol (Lausanne) 5:240
Davis PJ, Mousa SA, Cody V, Tang HY, Lin HY (2013) Small molecule hormone or hormone-like ligands of integrin αvβ3: implications for cancer cell behavior. Horm Cancer 4(6):335–342
Lin HY, Lansing L, Merillon JM, Davis FB, Tang HY, Shih A, Vitrac X, Krisa S, Keating T, Cao HJ, Bergh J, Quackenbush S, Davis PJ (2006) Integrin αvβ3 contains a receptor site for resveratrol. FASEB J 20(10):1742–1744
Lin HY, Chin YT, Yang YC, Lai HY, Wang-Peng J, Liu LF, Tang HY, Davis PJ (2016) Thyroid hormone, cancer, and apoptosis. Compr Physiol 6(3):1221–1237
Lee YS, Chin YT, Shih YJ, Nana AW, Chen YR, Wu HC, Yang YCSH, Lin HY, Davis PJ (2018) Thyroid hormone promotes β-catenin activation and cell proliferation in colorectal cancer. In: Horm Cancer, vol 9, pp 156–165
Lin HY, Chin YT, Nana AW, Shih YJ, Lai HY, Tang HY, Leinung M, Mousa SA, Davis PJ (2016) Actions of l-thyroxine and nano-diamino-tetrac (Nanotetrac) on PD-L1 in cancer cells. Steroids 114:59–67
Nana AW, Chin YT, Lin CY, Ho Y, Bennett JA, Shih YJ, Chen YR, Changou CA, Pedersen JZ, Incerpi S, Liu LF, Whang-Peng J, Fu E, Li WS, Mousa SA, Lin HY, Davis PJ (2018) Tetrac downregulates β-catenin and HMGA2 to promote the effect of resveratrol in colon cancer. Endocr Relat Cancer 25(3):279–293
Mousa, S.A., et al., Nano-diamino-tetrac (Nanotetrac), a thyroid hormone antagonist at integrin αvβ3, causes necrosis via anti-angiogenesis and induces apoptosis in human glioblastoma xenografts, in Late-Breaking Tumor Biology II (posters). 2016, Endocrine Society. p. LBSAT-03-LBSAT-03
Sudha T, Bharali DJ, Sell S, Darwish NHE, Davis PJ, Mousa SA (2017) Nanoparticulate tetrac inhibits growth and vascularity of glioblastoma xenografts. Horm Cancer 8(3):157–165
Moreno M, de Lange P, Lombardi A, Silvestri E, Lanni A, Goglia F (2008) Metabolic effects of thyroid hormone derivatives. Thyroid 18(2):239–253
Bharali DJ, Yalcin M, Davis PJ, Mousa SA (2013) Tetraiodothyroacetic acid-conjugated PLGA nanoparticles: a nanomedicine approach to treat drug-resistant breast cancer. Nanomedicine (Lond) 8(12):1943–1954
Lee YS, Chin YT, Yang YCSH, Wei PL, Wu HC, Shih A, Lu YT, Pedersen JZ, Incerpi S, Liu LF, Lin HY, Davis PJ (2016) The combination of tetraiodothyroacetic acid and cetuximab inhibits cell proliferation in colorectal cancers with different K-ras status. Steroids 111:63–70
Chin YT, Yang SH, Chang TC, Changou CA, Lai HY, Fu E, HuangFu W, Davis PJ, Lin HY, Liu LF (2015) Mechanisms of dihydrotestosterone action on resveratrol-induced anti-proliferation in breast cancer cells with different ERα status. Oncotarget 6(34):35866–35879
Lin HY, Tang HY, Davis FB, Davis PJ (2011) Resveratrol and apoptosis. Ann N Y Acad Sci 1215:79–88
Lin HY, Glinsky GV, Mousa SA, Davis PJ (2015) Thyroid hormone and anti-apoptosis in tumor cells. Oncotarget 6(17):14735–14743
Lin HY, Tang HY, Shih A, Keating T, Cao G, Davis PJ, Davis FB (2007) Thyroid hormone is a MAPK-dependent growth factor for thyroid cancer cells and is anti-apoptotic. Steroids 72(2):180–187
Yalcin M, Bharali DJ, Lansing L, Dyskin E, Mousa SS, Hercbergs A, Davis FB, Davis PJ, Mousa SA (2009) Tetraidothyroacetic acid (tetrac) and tetrac nanoparticles inhibit growth of human renal cell carcinoma xenografts. Anticancer Res 29(10):3825–3831
Nakano K, Bálint É, Ashcroft M, Vousden KH (2000) A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene 19(37):4283–4289
Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y (2000) A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404(6773):42–49
Yoshida Y, Tsunoda T, Doi K, Tanaka Y, Fujimoto T, Machida T, Ota T, Koyanagi M, Takashima Y, Sasazuki T, Kuroki M, Iwasaki A, Shirasawa S (2011) KRAS-mediated up-regulation of RRM2 expression is essential for the proliferation of colorectal cancer cell lines. Anticancer Res 31(7):2535–2539
Amin A et al (2009) Overview of major classes of plant-derived anticancer drugs. Int J Biomed Sci 5(1):1–11
Gweon EJ, Kim SJ (2013) Resveratrol induces MMP-9 and cell migration via the p38 kinase and PI-3K pathways in HT1080 human fibrosarcoma cells. Oncol Rep 29(2):826–834
Kim M, Murakami A, Kawabata K, Ohigashi H (2005) (-)-Epigallocatechin-3-gallate promotes pro-matrix metalloproteinase-7 production via activation of the JNK1/2 pathway in HT-29 human colorectal cancer cells. Carcinogenesis 26(9):1553–1562
Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW (1997) Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 275(5307):1787–1790
The Cancer Genome Atlas, N (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330–337
Yanamoto S, Kawasaki G, Yamada SI, Yoshitomi I, Yoshida H, Mizuno A (2009) Ribonucleotide reductase small subunit p53R2 promotes oral cancer invasion via the E-cadherin/β-catenin pathway. Oral Oncol 45(6):521–525
Lin HY, Tang HY, Keating T, Wu YH, Shih A, Hammond D, Sun M, Hercbergs A, Davis FB, Davis PJ (2008) Resveratrol is pro-apoptotic and thyroid hormone is anti-apoptotic in glioma cells: both actions are integrin and ERK mediated. Carcinogenesis 29(1):62–69
Chin YT, Wei PL, Ho Y, Nana AW, Changou CA, Chen YR, Yang YCSH, Hsieh MT, Hercbergs A, Davis PJ, Shih YJ, Lin HY (2018) Thyroxine inhibits resveratrol-caused apoptosis by PD-L1 in ovarian cancer cells. Endocr Relat Cancer 25(5):533–545
Davis PJ, Lin HY, Sudha T, Yalcin M, Tang HY, Hercbergs A, Leith JT, Luidens MK, Ashur-Fabian O, Incerpi S, Mousa SA (2014) Nanotetrac targets integrin αvβ3 on tumor cells to disorder cell defense pathways and block angiogenesis. Onco Targets Ther 7:1619–1624
Funding
This work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST104-2320-B-038-009; MOST105-2320-B-038-006; MOST104-2314-B-038-046-MY3).
Author information
Authors and Affiliations
Contributions
Concept and design of experiments: AWN and HYL
Acquisition of data: AWN, YTC, YJS, YCSHY, and YRC
Analysis and interpretation of data: AWN, YTC, TMC, YCSHY, AH, SI, and HYL
Writing and review of the manuscript: AWN, HYL, and PJD
Supervision of the work: HYL, SYW, WSL, YH, YML, YRL, JZP, SI, AH, LFL, JWP, and PJD
Corresponding author
Ethics declarations
Conflict of Interest
Co-author Paul J. Davis is co-inventor of Nanotetrac, a nanoparticulate analog of tetrac. The other authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Nana, A.W., Wu, S.Y., Yang, YC.S. et al. Nano-Diamino-Tetrac (NDAT) Enhances Resveratrol-Induced Antiproliferation by Action on the RRM2 Pathway in Colorectal Cancers. HORM CANC 9, 349–360 (2018). https://doi.org/10.1007/s12672-018-0334-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12672-018-0334-9