Abstract
Fungal pathogens are a major health issue that threatens the era of antifungal drugs commonly used in the treatment of infections. An effective approach of biosynthetic nanoparticles can be used as antifungal agents owing to their intrinsic features such as their simplicity, non-toxic, and physicochemical properties. Therefore, this study was aimed to molecularly ascertain Aspergillus species known to cause aspergillosis and investigate the potency of zinc oxide:gold nanoparticles (ZnO:Au NPs) against the fungal pathogens. Two Aspergillus strains retrieved with potato dextrose agar (PDA) culture media from commercial food products in South Africa were molecularly identified using calmodulin (CaM) gene region. DNA sequence phylogeny of the gene showed that the strains were A. niger and A. candidus. ZnO:Au (1%) NPs were synthesised and characterised using X-ray diffraction (XRD), scanning electron microscopy (SEM), and X-ray photoelectron spectroscopy (XPS). Two distinct peak plasmon bands for ZnO and ZnO:Au NPs were 390 nm and 565 nm, respectively. FE-SEM images demonstrated the presence of Au on the surface of ZnO nanoparticles in the ZnO:Au nanocomposites. The ZnO:Au NPs antifungal activity of 10 µg/mL and 50 µg/mL concentrations were evaluated against the two Aspergillus spp. ZnO:Au NPs at 50 µg/mL exhibited a maximum antifungal activity against A. candidus and A. niger, with zones of inhibition (ZoI) of 31.2 ± 0.15 mm and 25.0 ± 0.06, respectively. When the ZoI was observed by SEM, major morphological damages on the conidia were observed for both strains, indicating that the antifungal activity may have been enhanced by the ZnO:Au NPs. Therefore, due to these outstanding properties, ZnO:Au NPs can be utilised as potential antifungal agents to inhibit the proliferation of fungal pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Aspergillus is a ubiquitous, filamentous, mycotoxigenic fungal genus where few species (spp.) such as A. flavus, A. fumigatus, A. niger, and A. candidus are termed environmental opportunistic pathogens [1]. These species can also be harmful to humans, depending on the immune systems at the time of spore inoculation or inhalation, causing 90–100% infections, such as invasive aspergillosis (IA) [2], with a high mortality rate in immunocompromised and critically ill individuals and thus can spread locally or disseminate to distant sites.
Failure to receive early diagnosis contributes to poor prognosis of IA by delaying the start of antifungal therapy [3, 4]. Thus, making a conclusive diagnosis of fungal infections, particularly among immunocompromised hosts, remains an ongoing challenge. In addition, if there are any signs and symptoms of fungal infections, they are commonly not specific [5]. Culture-based diagnosis may lead to the identification of etiological agent, and the potential to be a time-consuming process. Additionally, the sample’s sensitivity may be affected by the site of collection, and contamination may go undetected [6].
Over the last years, clinical drug-resistant Aspergillus spp. have emerged as a result of misuse of existing antifungal drugs such as fluconazole, azoles, echinocandins, and polyenes [7, 8]. For example, azole resistance is rare and is caused by a change in the ergosterol content or the synthesis of other sterol types that form the membranes of fungal cells [2]. This mechanism may explain the resistance without compromising normal cell function. Nonetheless, studies have shown that azole drug resistance has severe clinical consequences with patients on drug-resistant aspergillosis exhibiting 25% increase in mortality [9]. However, recent studies propose using metallic nanoparticles for the management of fungal infections caused by Aspergillus sp. [10].
In recent years, nanotechnology has gained traction in research, and the application of nanoparticles (NPs) has been essential to the growth of this field in medicine and diagnostics, largely displacing conventional treatment approaches [11, 12]. Biosynthetic NPs offer several advantageous features depending on the metallic element, size, and shape of the particles, and less toxic, as well as exceptional catalytic and physicochemical properties [13, 14, 15, 16, 17, 18, 19, 20]. In particular, ZnO:Au NPs are well-known for their antifungal properties, with strong efficacy against the growth of A. niger [21] and A. candidus [22]. Essentially, resistance of Aspergillus sp. to routine antifungal treatment poses a major challenge to the management of infection; hence, the use of NPs as antifungal agents could help improve the management of Aspergillus infections.
In this present study, ZnO:Au NPs were synthesised and employed as antifungal agents against the molecularly identified strains of A. niger and A. candidus known to cause fungal infections. Herein, the two fungal strains were retrieved from commercial food products in South Africa using PDA culture media. Then, calmodulin (CaM) gene region was used for molecular identification. We hypothesised that the characterised ZnO:Au NPs, visualised in XRD, SEM, and XPS, could change the cell wall structure of the fungal strains through forming pores, possibly improving the sensitivity of the strains.
2 Materials and Methods
2.1 Materials
Zinc acetate dihydrate (CH3 COO)2 Zn·2H2O, 98.5%, SRL), methanol (99.8%, SRL), HAuCl4 3H2O ≥ 99.0%, Sigma Aldrich), sodium hydroxide (NaOH, 98%, SRL), methylene blue (Alfa Aesar C16H18CIN3S), and DI water (HPLC grade, Merck) were used.
2.2 Fungal Isolation
This procedure was performed according to the methods of Achilonu et al. [23, 24, 25]. Cereal grains from commercial food products in South Africa were surface sterilised for 30 s using 70% ethanol, rinsed with sterilised water (distilled water, dH2O), and dried for 5 min on a sterile bench. The grains were then transferred to a 90-mm Petri dish (Lasec, Bloemfontein, South Africa) containing potato dextrose agar (PDA: potato 200 g, dextrose 20 g, agar 16 g/L) (Merck-Millipore, Pretoria, South Africa) amended with chloramphenicol (0.01 g/L) (Biologica Pharmaceuticals, Pretoria). The Petri dishes were incubated for 7 days at 25 ±1 °C under 12-h alternating cycles of near-ultraviolet (NUV) (360-nm wavelength) light and darkness. Aspergillus cultures that developed on the original plates were moved to new PDA plates and classified into morphotypes according to variations in colour and texture [26]. Labcon LTGC-M40 incubator (Labcon, Gauteng, South Africa) was used to incubate the Petri dishes at 25 ±1 °C for 7 days under 12-h alternating cycles of near-ultraviolet (NUV) (360-nm wavelength) light and darkness. Pure Aspergillus cultures were obtained by performing a single hyphal tip removal and plated onto two distinct PDA media Petri dishes as A. niger (CGJM4174) and A. candidus (CGJM4177) under incubator conditions mentioned above and thus were resistant to propiconazole (azole) fungicide. Cultures of each strain were inoculated in 45 mL potato dextrose broth (PDB) media and placed in a shake-incubator (Apex-Scientific, Gauteng, South Africa), at a speed of 25 rpm for 14 days. Spore concentrations were adjusted to (1 × 106 spores/mL) using a hemocytometer [27], corresponding to the standardised number of viable spore counts per grid required for filamentous fungi.
2.3 Molecular Identification of Aspergillus Species
Genomic DNA (gDNA) was extracted from the mycelia (100 mg) using the ZR Quick-DNA Fungal/Bacterial Microprep™ Kit (Zymo Research, CA, USA). The DNA concentration and purity were determined with a NanoDrop Lite ND-2000 spectrophotometer (Thermo Fisher Scientific, MA, USA).
Polymerase chain reaction (PCR) was performed on the extracted gDNA using CaM gene primers (CMD5: 5′-CCG AGT ACA AGG ARG CCT TC-3′ and CMD6: 5′- CCG ATR GAG GTC ATR ACG TGG-3′) [28], according to the modified method of [23]. The PCR reaction (25 µL) consisted of 50 ng of template DNA, 0.3 µM of each primer, 2.5 mM MgCl2, 0.3 mM of each dNTP, and 1 U KAPA HiFi HotStart DNA Polymerase (Kapa Biosystems-Roche, Basel, Switzerland). T100TM Thermal Cycler conditions (Bio-Rad, Hercules, CA, USA) for PCR amplification entailed an initial denaturation step of 3 min at 95 °C followed by 25 cycles of 20 s at 98 °C, 15 s annealing step at 62.5 °C, 60 s at 72 °C, and a final elongation step of 3 min at 72 °C. The PCR amplicons were stained with GelRed nucleic acid stain (Thermo Fisher Scientific) and were examined using 2% agarose gel electrophoresis, visualised with a UV light ‘Gel Doc EZ Gel Documentation System’ (Bio-Rad).
The purified PCR amplicons for all the genes were sequenced in both primer directions using the BigDye Terminator v.3.1 kit (Applied Biosystems, CA, USA). Reactions contained 1 × BigDye Terminator Premix, 3.2 µM PCR primer, 5 × sequencing buffer, and 3 µL PCR amplicon, in a total volume of 10 µL. The conditions for the thermal cycler consisted of an initial denaturation step at 96 C for 1 min, followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min. Cycle sequencing products were purified using the ZR DNA Sequencing Clean-UpTM Kit (Zymo Research). ABI 3500xl Genetic Analyzer (Applied Biosystems) was used to analyse the purified sequencing products in accordance with industry standard procedures.
The phylogenetic analysis was performed using a modified method of [23, 24]. The generated A. niger and A. candidus DNA sequences and CaM were aligned with the reference sequence dataset with accession numbers (Supplementary Table S1) using the ClustalW parameter on Geneious Prime v. 2023.2 [29]. Maximum likelihood (ML) phylogenetic tree with 1000 bootstrap replicates was constructed with the appropriate model obtained by MEGA X. v.10.1 [30].
2.4 Synthesis of Pure ZnO and ZnO:Au NPs
Sol-gel approach was chosen for the synthesis of nanoparticles due to its adaptability, precision, and ability to produce nanoparticles with controlled size, shape, and composition. The synthesis of pure ZnO and ZnO:Au NPs was achieved using the sol–gel technique. After combining 40 mL of methanol with 3 g of zinc acetate dihydrate, the mixture was stirred at room temperature for 5 min. This led to the preparation of a zinc acetate solution. Another solution was prepared by mixing 10 mL of methanol with 15 g of sodium hydroxide. The two solutions were then combined with continuous stirring for a period of up to 10–15 min until white precipitates formed. After filtering the resulting precipitate, it was washed many times with methanol and allowed to dry at room temperature overnight. The samples were then dried in an oven at 90 °C for 24 h. The samples were further calcined in a muffle furnace at 550 °C for 2 h to promote the nucleation and development of pure ZnO nanoparticles.
The entire chemical reaction to form ZnO nano-powder using sodium hydroxide as a solvent is given as follows [31, 32]:
For doping, a solution containing 1 mol% of HAuCl4·3H2O was then added to previous prepared zinc acetate solution to create a uniform combination throughout. Another solution was prepared by mixing 10 mL of methanol with 15 g of sodium hydroxide. The two solutions were then combined with continuous stirring for a period of up to 10–15 min until white precipitates formed. After filtering the resulting precipitate, it was washed many times with methanol and allowed to dry at room temperature for a whole night. The samples were then dried in an oven at 90 °C for 24 h. The samples were further calcined in a muffle furnace at 550 °C for 2 h to promote the nucleation and development of ZnO:Au nanoparticles. To check the composition and properties of the synthesised nanoparticles, various approaches were used for the characterisation.
2.5 Material Testing Procedure
2.5.1 X-ray Diffraction Analysis
The prepared ZnO and ZnO:Au NPs powders were subjected to a characterisation process. Herein, the phase identification of the prepared samples was examined using X-ray powder diffraction (a D8 Advance X-ray diffractometer) (Bruker, Germany) with CuKα radiation (λ = 0.15408 nm) between 10 and 90° scan range and a voltage of 40 kV or 40 mA. The scanning range ranged from 20 to 80° and enabled a comprehensive examination of the crystalline composition of the powders.
2.5.2 UV-Vis Analysis
The obtained ZnO and ZnO:Au NPs powders were subjected to UV-Vis spectroscopic measurements after the annealing process. These measurements were performed in the wavelength range of 250 to 800 nm using a Lambda 950 UV-Vis-NIR spectrophotometer equipped with a PerkinElmer integrating sphere. The tests were carried out in the physics department of the University of the Free State in Bloemfontein, South Africa.
2.5.3 Scanning Electron Microscopy of ZnO and ZnO:Au Nanoparticles
Imaging of ZnO and ZnO:Au NPs was achieved by using field emission scanning electron microscopy (FE-SEM) with a model JSM-7800 F from JEOL. To create high-resolution scanning electron micrographs, the microscope is equipped with a Schottky field emission gun (T-FEG) and super hybrid lenses. These lenses combine a magnetic field lens with an electrostatic lens. For this experiment, a scanning electron microscope (SEM) photograph was taken using an electron beam with a power of 5.0 kV and a range of ×5000 to ×30,000 magnification. The electron beam voltage remained constant at 10 keV throughout the study, ensuring the accuracy of the data and the highest possible resolution. To accurately analyse the elemental composition of the samples, a sophisticated energy-dispersive X-ray spectrometer (EDS) was also used. This EDS was easily connected to scanning electron microscope (SEM) equipment.
2.5.4 X-ray Photoelectron Spectroscopy Analysis
An ultra-high vacuum (UHV) workspace was used to conduct X-ray photoelectron spectroscopy (XPS) experiments. The base pressure was kept below 2 × 10−9 Torr to ensure the most favourable conditions for analysis. The tests were carried out with great care and attention to detail using a PHI 5400. To ensure accuracy, the instrument’s working function needed to be calibrated. This was achieved by setting the binding energy (BE) of the Au 4f7/2 line for metallic gold to 83.96 eV and changing the spectrometer dispersion to give a BE of 932.67 eV for the Cu 2p3/2 line (which stands for metallic copper). To ensure the consistency of all measurements, a photoelectron recording angle of 45 °C was maintained. It was determined that survey scans should be performed with a pass energy of 178.95 eV, with energy steps of 0.25 eV, and a dwell time of 100 ms. To perform high-resolution scans, a pass energy of 44.75 eV and a step size of 0.125 eV were used. The charge correction was performed concerning the binding energy of the C 1s random energy envelope, which was 284.8 eV [19, 33, 34].
2.6 In Vitro Antifungal Activity of ZnO:Au Nanoparticles
The antifungal activity of ZnO:Au NPs was investigated against the two fungal strains, A. niger (CGJM4174) and A. candidus (CGJM4177), using a well-diffusion technique [4]. Conidial suspensions of each pathogen grown in 5 mL potato dextrose broth (PDB) media were mixed with potato dextrose agar (PDA) medium (50 °C) and poured in separate 90-mm Petri plates (Lasec, Bloemfontein, South Africa). The amended plates were allowed to solidify under a laminar air-flow system. A sterile plug borer of 5 mm was used to bore wells in the PDA plates, and 50 µL of each AuNP at varying concentrations (10 and 50 µg/mL in sterile dH2O) was added to each amended inoculum. For every plate, 50 µL of dH2O (negative control) and 5% sodium hypochlorite (NaOCl) as the positive control were added in separate wells. The assay was performed in triplicate and the plates were incubated for 5 days of alternating 12-h cycles of near-ultraviolet (NUV) light and darkness at 25 ± 1 °C in a Labcon LTGC-M40 incubator (Labcon, Gauteng, South Africa).
ImageJ v. 1.53 software [35] was used to measure the diameter (mm) of the zone where the ZnO:Au NPs and the positive controls inhibited the fungal growth. The scale standard of the images was established by calibrating two measurements of the plate’s orthogonal diameter (200 mm). After determining the inhibition zone diameters at 0° and 90° measurements, the mean diameter values were applied. Standardised sizes of the inhibition zones were obtained by subtracting the values from the respective wells from the ZnO:Au NPs zone of inhibition (ZoI).
2.7 Scanning Electron Microscopy on Inhibited Aspergillus Species
Scanning electron microscopy (SEM) was used to evaluate the treated plates (10 µg/mL) for each fungal strain exhibiting zones of inhibition. From the plates, small blocks (5 mm) of agar containing the edges of the zones of inhibition and the untreated (negative control) mycelial growth zones were aseptically cut out and the samples were fixed according to the methods of Achilonu et al. [4]. Herein, the samples were fixed in 3% glutaraldehyde (Sigma Aldrich, St. Louis, Missouri, USA), rinsed in 0.1 M (pH 7.0) sodium phosphate buffer solution for 3 h, and postfixed in 1% osmium tetroxide (OsO4) for 1 h. Samples were placed on 0.2-μm polycarbonate membrane filters and dehydrated in a graded ethanol series (50%, 70%, and 95%) for 20 min in each phase, followed by two changes in 100% for 1 h in each phase. Samples were dried using a critical point dryer (Tousimis, Maryland, USA), mounted on stubs (Cambridge pin type, 10 mm) using double-sided carbon tape and gold coated (± 60 nm) with a Bio-Rad sputter coater (Bio-Rad, Oxfordshire, UK). Finally, the samples were examined and analysed with a JSM-7800 F Extreme-resolution Analytical Field Emission Scanning Electron Microscope (JEOL Ltd, Tokyo, Japan) at the Centre for Microscopy, University of the Free State, South Africa.
2.8 Statistical Analysis
All the antifungal activity data (mm) were performed in triplicate and represented as means ± standard deviations (SD). The means were statistically compared using two-way ANOVA using R version 4.1.0 [36] within R-Studio v. 1.3.959 [37], and Fisher’s LSD test (p = 0.05) function from the ‘agricolae’ [38] and ‘doebioresearch’ (Analysis of Factorial Randomised Block Design for 3 factors) [39] packages were used to determine the significant level.
3 Results and Discussion
3.1 DNA Sequence and Phylogenetic Analysis
The two fungal strains were successfully amplified using CaM genes, with amplicon sizes of approximately 740 base pairs (Supplementary Fig. S1). Fungal strain sequences were submitted to the National Center for Biotechnology Information (NCBI) GenBank, and queried isolates had over 96% similar identity to the GenBank reference sequences with their accession numbers. The ML phylogenetic tree consisting of 17 in-group Aspergillus sp. and 1 out-group taxa of Fusarium proliferatum showed two different clustering patterns that are very identical to the GenBank reference fungal sequences (Fig. 1). In addition, A. niger and A. candidus clustered distinctly with bootstrap values of 85% and 94%, respectively. In addition, the strains in the cluster are displayed in bold black colour, while the reference GenBank sequences are displayed with their accession numbers. Aspergillus species are ubiquitous and important human pathogens causing various kinds of disease [2]. Rapid and accurate identification of potential fungal pathogens at the species is often critical to disease surveillance [28, 40] and the implementation of a disease control strategy such as nanoparticle application [21]. Hence, this study established the presence of A. niger and A. candidus in commercial food products based on the molecular assay, which includes characterisation of CaM gene. These outcomes are to be expected given that these microbes are ubiquitous in nature and are predominant in the environment [41, 42]. Therefore, the occurrence of Aspergillus sp. in the current study has significant importance because these pathogens with mycotoxins have been associated with health deterioration or death in immunocompromised individuals [1, 43, 44].
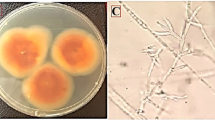
DNA-based (ML) phylogenetic tree of Aspergillus niger and Aspergillus candidus strains of CaM gene sequence. Bootstrap support values (1000 replicates) are indicated on the branch nodes below branches. Fusarium proliferatum was used as an outgroup and the two identified Aspergillus spp. are bolded in light-green frames. Macroscopic morphology of A. niger (a) and A. candidus (b) depicting the representative culture strain
3.2 Structural Study: X-ray Diffraction
The XRD patterns for both pure ZnO and Au-ZnO nanopowders in the diffraction angle range of 2\(\theta\) = 20–60° are shown in Fig. 2. In the pure ZnO sample, the observed peaks correspond to the hexagonal wurtzite structure of ZnO, as indicated by characteristic peaks with Miller indices of (100), (002), (101), and (102) [16, 19]. The XRD patterns of Au-doped ZnO, as shown in Fig. 2, also exhibit peaks indicative of the hexagonal ZnO phase. In addition, additional peaks related to the cubic structure of the Au phase were observed, and aligned, particularly at 111 and 200 [45]. The intensity of the Au peaks was observed at the Au 1% concentration (mol), suggesting the presence of stable metallic Au as a secondary crystal phase in the ZnO nanopowders. The presence of Au peaks in the XRD patterns of Au-ZnO is consistent with the results of a previous report [45]. The significantly larger radius of Au3+ (0.126 nm) compared to Zn2+ (0.070 nm) is a possible reason for the presence of the secondary crystalline phase in the Au-ZnO samples. The difference between the radii of Au3+ and Zn2+ limits the solubility of Au in the Zn lattice site [18]. This may be due to the successful incorporation of dopants into ZnO and the presence of microstructural stresses due to defects in Au-ZnO. The crystallite sizes (D) of pure ZnO and ZnO:Au NPs were estimated from the FWHM of all peaks using Scherrer’s formula. The average crystallite sizes of the prepared samples were found to be 19 nm for pure ZnO. The doping of ZnO:Au (1 mol %) resulted in XRD peaks and thus average crystallite sizes of 28 nm. However, ZnO doped with 1 mol% Au showed narrower peaks than pure ZnO, indicating an increase in average crystallite size.
3.3 Optical Properties of ZnO:Au Nanoparticles
The UV–visible absorbance spectrum for both bare ZnO NPs and ZnO:Au nanostructures is shown in Fig. 3. The ZnO NPs exhibited a characteristic absorption band edge at 390 nm, which is attributed to the absorbance peak of pure ZnO [4, 26]. In the absorption spectra of Au-ZnO nanocomposites, two distinct peaks were observed: one in the UV region around 390 nm, attributed to the band edge absorption of ZnO, and the other in the visible region around 565 nm, attributed to the surface plasmon absorption of Au NPs [18]. The influence of the plasmon band is influenced by various factors, including particle size, charge density of Au NPs, interparticle spacing, and the surrounding dielectric medium [33]. The characteristic plasmon peak of Au-ZnO nanocomposites appeared broad in the visible region, indicating a wide particle size distribution. The bandgaps of the pure ZnO and doped sample with Au were found to be 3.16 eV and 3.13 eV (see SM-1).
3.4 Surface Morphology Analysis of ZnO:Au Nanoparticles
The surface morphology, elemental compositions, and micrographs of the pure ZnO and Au-doped ZnO nanostructures were observed using field emission-scanning electron microscopy (FES-EM) coupled with EDS (Fig. 4a–d). The FE-SEM images were analysed from the different scanned areas and showed almost flower-shaped aggregated clusters (Fig. 4). The irregular solid clusters were formed due to the slow rate of crystal growth at the applied temperature with the underutilised sol-gel technique. The surface directing or capping agents were not added during the nucleation and growth mechanisms of the particles, and this could be a reason for the clustering of ZnO particles. Similarly, agglomerated compact structures were observed for the different concentrations of Au-doped ZnO, which exhibited almost identical surface morphologies. However, the EDS profile only confirmed the presence of Zn and O elements, indicating a pure form of the ZnO sample (Fig. 4), while for the doped ZnO samples, the presence of Au was detected in the EDS spectra (Fig. 4d). Furthermore, the colour mapping of the Au-doped ZnO samples indicating the distribution of Au, Zn, and O individually, as shown in Fig. 5a–d.
3.5 X-ray Photoelectron Spectroscopy (XPS) Analysis
The XPS examination confirmed the presence of C, O, Au, and Zn peaks, as shown in Fig. 6a [18]. Binding energy calibration for charge correction in the high-resolution spectra was performed using C-1 s at a standard value of 284.8 eV [16, 17, 18, 19, 33, 34, 46, 47, 48, 49]. The high-resolution spectrum of C-1s for pure ZnO is shown in Fig. 6b. The spectrum was resolved into six Gaussian fits for the different chemical states of carbon. Peaks are assigned to metal carbide (283.6 eV), C-C (284.8 eV), C-sp3/C-N/defects (285.4 eV), and C-O (286 eV), C-HO (287.3 eV), and C = O/CO3 (289 eV). Figure 6c shows a representation of the core-level XPS spectrum of Zn 2p for pure ZnO. An examination of the Zn 2p3/2 envelope reveals three unique peaks, each of which is indicative of a different chemical state of zinc. Stoichiometric zinc oxide is responsible for the significant peak that was detected at 1021.6 eV, whereas Zn(OH)2 is responsible for another noticeable peak that was observed at 1022.6 eV. There is also a third peak that can be seen at 1019.8 eV, which corresponds to the surface defects [16]. In addition, the Zn LMM peaks were used to determine the oxidation state of zinc in pure ZnO samples. The Zn-LMM peak of pure ZnO was deconvoluted into three peaks, which can be seen in Fig. 6d: the primary peak, which appears at 988.6 eV and corresponds to ZnO; a conspicuous peak, which appears at 986.3 eV and is attributable to Zn(OH)2; and a third peak, which appears at 991.3 eV and is suggestive of Zn interstitials.
ZnO is doped with Au, as shown in Fig. 7, and a slight change in the binding energy of Zn2+ is observed. Fits corresponding to the Zn 2p3/2 peak for ZnO doped with Au were performed and are also shown in Fig. 7a. These peak placements are consistent with reports from other studies [16]. In the pure ZnO sample, the major peak in the spectrum, which was found at 988.6 eV, represents Zn2+. Similar results were achieved regarding the oxidation state of Zn in ZnO [16] that had been doped with varied concentrations of Au (Fig. 7b). Core binding energies of the Zn-3p and Au-4f electrons are shown in Fig. 7c. The Zn 3p binding energy for the Au-ZnO and ZnO samples is resolved into two peaks representing Zn 3p3/2 (88.1 eV), Zn 3p1/2 (91.1 eV), and Au 4f7/2 ( 82.90 eV), which thus corresponds to Au 4f5/2 (86.65 eV) as shown in Fig. 7c and other similar studies [18]. Notably, the position of the Zn-3p binding energy peak remains consistent across all samples, indicating an unchanged chemical state of Zn upon confinement of Au on the surface of ZnO. However, the binding energy of Au 4f is shifted to lower values compared to pure gold (4f7/2, 84.00 eV and 4f5/2, 87.71 eV) as shown in Fig. 7c, which corresponds to similar studies [46]. The first peak observed at 82.9 eV was assigned to pure Au; the other peak observed at 86.65 eV was assigned to Au3+. This shift is attributed to the formation of negatively charged Au nanoparticles without forming chemical bonds with ZnO. The Fermi level of Au (5.4 eV) and the donor level of ZnO are nearly equal, indicating the potential for electron transfer from ZnO to Au, which would raise the charge density on the surface of Au NPs [18]. .
3.6 In Vitro Antifungal Activity Efficacy of ZnO:Au NPs
The in vitro antifungal activity of ZnO:Au NPs showed a promising influence as an antifungal agent to the two fungal spp. with minimum inhibitory concentration (MIC) (Table 1; Fig. 8). At 50 µg/mL concentration, A. candidus is the most common fungal strain sensitive to ZnO:Au NPs (Fig. 8a) followed by A. niger (Fig. 8c), with 31.2 mm and 25.0 mm ZoI (Table 1), respectively. This indicates significant differences (p < 0.001) between the two fungal ZoI values (Table 2), while A. candidus (Fig. 8b) and A. niger (Fig. 8d) showed the least sensitivity, with 14.4 mm and 13.1 mm ZoI, respectively at 10 µg/mL dose of ZnO:Au NPs and differed significantly (p < 0.001). When compared to positive controls (5% NaOCl) and the ZnO:Au NPs treatment (Table 1), the negative controls (dH2O) for both strain treatments showed no antifungal effect with 0-mm ZoI and differed significantly (p < 0.001). The in vitro antifungal activity of ZnO:Au NPs showed varying levels of inhibitory effects on the two species of Aspergillus. Similar studies have shown that ZnO:Au NPs have great efficacy to inhibit the growth of other Aspergillus spp. such as A. flavus and A. fumigatus [10, 21], as well as other pathogenic fungi like Streptomyces cyaneus [50]. Nonetheless, our findings showed that the antifungal activity of ZnO:Au NPs caused the integrity of the cell membrane to be destroyed, which in turn caused distorted cells and cell death. These morphological alterations may be linked to disruptions in the morphogenesis and growth of Aspergillus sp., as previously reported [51, 52].
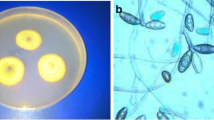
Antifungal activity studies of ZnO:Au NPs against Aspergillus niger (CGJM4174) and Aspergillus candidus (CGJM4177) strains with zone of inhibition (ZoI). Black arrow indicates antifungal activities caused by 50 and 10 µg/mL concentration of the ZnO:Au NPs, (+ve) denotes (5% NaOCl) positive control, and (−ve) (dH2O) negative control
3.7 Scanning Electron Microscopy on Aspergillus Species
The SEM images of the untreated (dH2O-negative control) and treated (ZnO:Au NPs) conidia are shown in Fig. 9a–d respectively. From Fig. 9a, we can observe that the cells of A. candidus are intact having a smooth surface. However, the conidia for the treated showed changes in cell morphology such as plasmolysis, distortions, and being squashed (Fig. 9b). Similarly to the untreated A. niger (Fig. 9c), the conidia were uncompromised with intact cells. On the contrary, there were cell shrinkage and collapsed or rough conidial surfaces for the treated A. niger conidia (Fig. 9d). The findings clearly showed a substantial morphological change as well as damage between the conidia that were treated and untreated. This could be the consequence of intracellular compound leakage from damaged cell membranes, which would completely disrupt the cells, as reported in similar studies using ZnO:Au NPs against species of Aspergillus [10, 53, 54].
Scanning electron microscopy (SEM) micrograph of Aspergillus niger (CGJM4174) and Aspergillus candidus (CGJM4177) hyphal morphological changes directly exposed to ZnO:Au NPs (10 µg/mL). a A. candidus conidia treated with dH2O ( − ve control). b A. candidus conidia treated with AuNPs. c A. niger conidia treated with dH2O (control). d A. niger conidia treated with ZnO:Au NPs. The green arrow denotes the smooth cell membrane of conidia. Red arrow depicts distorted or rough conidial surface. Scale bar = 1 and 10 μm
Based on our knowledge, there are hardly any reports on the mode of antifungal action of ZnO:Au NPs against A. niger and A. candidus, despite the fact that this precise mechanism is not fully understood. Nonetheless, gaining insight into the various potential modes of action of ZnO:Au NPs inhibitory effect could involve the combination of different routines of mechanisms [2]. These mechanisms enable nanomaterials to adhere to the cell membrane, altering their permeability and structure resulting in impaired membrane transport in the ABC transporter system (ATP binding cassette) and impeded ATP production [55]. Nanomaterial penetration of the cells inhibits the synthesis of ergosterol, ATP, and aminoacyl tRNA synthetase, leading to cell dysfunction and altered sterol metabolism, ultimately resulting in fungal cell leakage, plasmolysis, and cell lysis. Another route of mechanism involves interfering with related proteins to damage the signalling pathways, which prevents the cell from carrying out essential physiological functions [56]. Cells become inactive by impairing their ability to replicate DNA, oxidizing proteins, and lipids, leading to cellular toxicity [4, 51]. Nanomaterial mode of action in the cell causes the release of reactive oxygen species (ROS) and free radicals leading to oxidative stress which plays an important role in inducing apoptosis [2, 10, 21]. Furthermore, the antifungal mechanism of ZnO:Au NPs on the fungal cell membrane is probably due to the reaction with the medium components which promoted the discharge of Au+ ions [57, 58]. The formed Au+ ions may produce free radicals and induce intracellular oxidative stress to the fungal cells [59, 60]. This indicates that a variety of targets within the fungal cells are affected by the fungicidal effects of the ZnO:Au NPs agent. Therefore, in light of our findings, it is safe to propose that ZnO:Au NPs have strong antifungal properties which make them a good substitute agent in controlling antifungal drug-resistant pathogens [21]. This indicates that a variety of targets within the fungal cells are affected by the fungicidal effects of the ZnO:Au NPs agent. Therefore, in light of our findings, it is safe to propose that ZnO:Au NPs have strong antifungal properties which make them a good substitute agent in controlling antifungal drug-resistant pathogens [21].
4 Conclusion
The two Aspergillus spp. were successfully identified using DNA sequence phylogeny, thereby validating the morphotype characteristic cultures. However, Au and Zn have long been utilised for their antifungal properties and have proven to be a great option. This research showed the low-cost, feasible, and eco-friendly process, and the successful antifungal efficacy of ZnO:Au NPs against A. niger and A. candidus strains known to be multidrug resistant. The XRD study undeniably confirms the presence of hexagonal wurtzite structures characteristic of ZnO, which agrees well with the XPS results found. Furthermore, SEM and EDS studies provide evidence that ZnO possesses morphologies that are free of detectable impurities. This further supports the consistency found in the results of XPS and UV-visible absorption as well as XRD-based research findings. Based on the experimental findings, the two Aspergillus spp. were sufficiently restrained and attacked by the novel synthetic type NPs containing Au, as indicated by their low ZoI and MIC. These NPs contained a bell shape in the UV-Vis analysis, indicating that the ZnO:Au NPs were spherical and monodispersed as supported by the FESEM-EDX analysis. It is noteworthy that the primary factors affecting ZnO:Au NPs’ ability to suppress these fungal diseases were their small crystal and particle sizes. Based on the encouraging outcomes, ZnO:Au NPs have the potential to function as a potent antifungal agent that can be employed in the fight against fungal infections.
Data Availability
No datasets were generated or analysed during the current study.
References
Navale, V., Vamkudoth, K. R., Ajmera, S., & Dhuri, V. (2021). Aspergillus derived mycotoxins in food and the environment: Prevalence, detection, and toxicity. Toxicology Reports, 8, 1008–1030. https://doi.org/10.1016/j.toxrep.2021.04.013
Achilonu, C. C., Davies, A., Kanu, O. O., Noel, C. B., & Oladele, R. (2023). Recent advances and future perspectives in mitigating invasive antifungal-resistant pathogen Aspergillus fumigatus in Africa. Current Treatment Options in Infectious Diseases, 15(4), 1–20. https://doi.org/10.1007/s40506-023-00269-4
Kanj, S. S., Omrani, A. S., Al-Abdely, H. M., Subhi, A., Fakih, R. E., Abosoudah, I., & Dimopoulos, G. (2022). Survival outcome of empirical antifungal therapy and the value of early initiation: A review of the last decade. Journal of Fungi, 8(11), 1146. https://doi.org/10.3390/jof8111146
Achilonu, C. C., Gryzenhout, M., Marais, G. J., Johar, D., Ghosh, S., & Hassanin, S. O. (2024). Antifungal activity of Carya illinoinensis extracts against Alternaria alternata pathogen and their cytotoxicity effects on HEK-293T cells: HPLC analysis of bioactive compounds. Discover Applied Sciences, 6(2), 67. https://doi.org/10.1007/s42452-024-05721-8
Sanguinetti, M., Posteraro, B., Beigelman-Aubry, C., Lamoth, F., Dunet, V., Slavin, M., & Richardson, M. D. (2019). Diagnosis and treatment of invasive fungal infections: Looking ahead. Journal of Antimicrobial Chemotherapy, 74(Supplement_2), ii27–ii37. https://doi.org/10.1093/jac/dkz041
Zhang, S. X., Babady, N. E., Hanson, K. E., Harrington, A. T., Larkin, P. M. K., Leal, S. M., on behalf of the Fungal Diagnostics Laboratories Consortium (FDLC). (2021). Recognition of diagnostic gaps for laboratory diagnosis of fungal diseases: Expert opinion from the Fungal Diagnostics Laboratories Consortium (FDLC). Journal of Clinical Microbiology, 59(7). https://doi.org/10.1128/jcm.01784-20
Wiederhold, N. P. (2022). Emerging fungal infections: New species, new names, and antifungal resistance. Clinical Chemistry, 68(1), 90. https://doi.org/10.1093/CLINCHEM/HVAB217
Campbell, C. A., Osaigbovo, I. I., & Oladele, R. O. (2021). Triazole susceptibility of Aspergillus species: Environmental survey in Lagos, Nigeria and review of the rest of Africa. Therapeutic Advances in Infectious Disease, 8, 20499361211044330. https://doi.org/10.1177/20499361211044330
Lestrade, P. P., Bentvelsen, R. G., Schauwvlieghe, A. F. A. D., Schalekamp, S., Van Der Velden, W. J. F. M., Kuiper, E. J., & Verweij, P. E. (2019). Voriconazole resistance and mortality in invasive aspergillosis: A multicenter retrospective cohort study. Clinical Infectious Diseases, 68(9), 1463–1471. https://doi.org/10.1093/CID/CIY859
Elegbede, J. A., Lateef, A., Azeez, M. A., Asafa, T. B., Yekeen, T. A., Oladipo, I. C., & Gueguim-Kana, E. B. (2020). Biofabrication of gold nanoparticles using xylanases through valorization of corncob by Aspergillus niger and Trichoderma longibrachiatum: Antimicrobial, antioxidant, anticoagulant and thrombolytic activities. Waste and Biomass Valorization, 11(3), 781–791. https://doi.org/10.1007/s12649-018-0540-2
Haleem, A., Javaid, M., Singh, R. P., Rab, S., & Suman, R. (2023). Applications of nanotechnology in medical field: A brief review. Global Health Journal, 7(2), 70–77. https://doi.org/10.1016/j.glohj.2023.02.008
Joseph, T. M., Kar Mahapatra, D., Esmaeili, A., Piszczyk, Ł, Hasanin, M. S., Kattali, M., & Thomas, S. (2023). Nanoparticles: Taking a unique position in medicine. Nanomaterials, 13(3), 574. https://doi.org/10.3390/nano13030574
Chen, Y., Zeng, D., Zhang, K., Lu, A., Wang, L., & Peng, D. L. (2014). Au–ZnO hybrid nanoflowers, nanomultipods and nanopyramids: One-pot reaction synthesis and photocatalytic properties. Nanoscale, 6(2), 874–881. https://doi.org/10.1039/C3NR04558G
Gogurla, N., Sinha, A. K., Santra, S., Manna, S., & Ray, S. K. (2014). Multifunctional Au-ZnO plasmonic nanostructures for enhanced UV photodetector and room temperature NO sensing devices. Scientific Reports, 4(1), 6483. https://doi.org/10.1038/srep06483
Hou, X., & Wang, L. (2014). Controllable fabrication and photocatalysis of ZnO/Au nanohybrids via regenerative ion exchange and reduction cycles. RSC Advances, 4(100), 56945–56951. https://doi.org/10.1039/C4RA10785C
Kumar, P., Kumar, A., Rizvi, M. A., Moosvi, S. K., Krishnan, V., Duvenhage, M. M., & Swart, H. C. (2020). Surface, optical and photocatalytic properties of Rb doped ZnO nanoparticles. Applied Surface Science, 514, 145930. https://doi.org/10.1016/j.apsusc.2020.145930
Kumar, P., Mathpal, M. C., Inwati, G. K., Ghosh, S., Kumar, V., Roos, W. D., & Swart, H. C. (2020). Optical and surface properties of Zn doped CdO nanorods and antimicrobial applications. Colloids and Surfaces A: Physicochemical and Engineering Aspects, 605. https://doi.org/10.1016/j.colsurfa.2020.125369.
Kumar, P., Chandra Mathpal, M., Jagannath, G., Prakash, J., Maze, J. R., Roos, W. D., & Swart, H. C. (2021). Optical limiting applications of resonating plasmonic Au nanoparticles in a dielectric glass medium. Nanotechnology, 32(34). https://doi.org/10.1088/1361-6528/abfee6
Kumar Inwati, G., Kumar, P., Roos, W. D., & Swart, H. C. (2020). Thermally induced structural metamorphosis of ZnO:Rb nanostructures for antibacterial impacts. Colloids and Surfaces B Biointerfaces, 188, 110821. https://doi.org/10.1016/j.colsurfb.2020.110821
Nagajyothi, P. C., Lim, H., Shim, J., & Rawal, S. B. (2016). Au nanoparticles supported nanoporous ZnO sphere for enhanced photocatalytic activity under UV-light irradiation. Journal of Cluster Science, 27(4), 1159–1170. https://doi.org/10.1007/s10876-016-0980-4
Almansob, A., Bahkali, A. H., & Ameen, F. (2022). Efficacy of gold nanoparticles against drug-resistant nosocomial fungal pathogens and their extracellular enzymes: Resistance profiling towards established antifungal agents. Nanomaterials, 12(5), 836. https://doi.org/10.3390/NANO12050814
Vala, A. K. (2014). Investigations on gold nanoparticles biosynthesis potential of marine derived fungi. BioTechnology: An Indian Journal, 9(5):206–209.
Achilonu, C. C., Marais, G. J., Ghosh, S., & Gryzenhout, M. (2023). Multigene phylogeny and pathogenicity trials revealed Alternaria alternata as the causal agent of black spot disease and seedling wilt of pecan (Carya illinoinensis) in South Africa. Pathogens, 12(5), 672–691. https://doi.org/10.3390/pathogens12050672
Achilonu, C. C., Gryzenhout, M., Marais, G. J., & Ghosh, S. (2023). Differential detection of Alternaria alternata haplotypes isolated from Carya illinoinensis using PCR-RFLP analysis of Alt a1 gene region. Genes, 14(5), 1115–1129. https://doi.org/10.3390/genes14051115
Achilonu, C. C., Gryzenhout, M., Marais, G. J., Madisha, M. T., & Ghosh, S. (2023). Random amplified microsatellites (RAMS) analysis ascertains genetic variation of Alternaria alternata causing black spot disease on Carya illinoinensis in South Africa. Frontiers in Genetics, 14. Retrieved from https://www.frontiersin.org/articles/https://doi.org/10.3389/fgene.2023.1213102.
Gamal, A., Fikry, E., Tawfeek, N., El-Shafae, A. M., El-Sayed, A. S. A., & El-Domiaty, M. M. (2024). Production and bioprocessing of taxol from Aspergillus niger, an endophyte of encephalartos whitelockii, with a plausible biosynthetic stability: Antiproliferative activity and cell cycle analysis. Microbial Cell Factories, 23(1), 78. https://doi.org/10.1186/s12934-024-02356-7
Camacho-Fernández, C., Hervás, D., Rivas-Sendra, A., Marín, M. P., & Seguí-Simarro, J. M. (2018). Comparison of six different methods to calculate cell densities. Plant Methods, 14(1), 30–45. https://doi.org/10.1186/S13007-018-0297-4
Alshehri, B., & Palanisamy, M. (2020). Evaluation of molecular identification of Aspergillus species causing fungal keratitis. Saudi Journal of Biological Sciences, 27(2), 756. https://doi.org/10.1016/J.SJBS.2019.12.030
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., … Drummond, A. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549. https://doi.org/10.1093/molbev/msy096
Haque, M. J., Bellah, M. M., Hassan, M. R., & Rahman, S. (2020). Synthesis of ZnO nanoparticles by two different methods & comparison of their structural, antibacterial, photocatalytic and optical properties. Nano Express, 1(1), 010007. https://doi.org/10.1088/2632-959X/ab7a43
Hasnidawani, J. N., Azlina, H. N., Norita, H., Bonnia, N. N., Ratim, S., & Ali, E. S. (2016). Synthesis of ZnO nanostructures using sol-gel method. Procedia Chemistry, 19, 211–216. https://doi.org/10.1016/j.proche.2016.03.095
Kumar, P., Mathpal, M. C., Prakash, J., Hamad, S., Rao, S. V., Viljoen, B. C., … Swart, H. C. (2019). Study of tunable plasmonic, photoluminescence, and nonlinear optical behavior of Ag nanoclusters embedded in a glass matrix for multifunctional applications. Physica Status Solidi (A), 216(4), 1800768. https://doi.org/10.1002/pssa.201800768
Kumar, P., Mathpal, M. C., Hamad, S., Rao, S. V., Neethling, J. H., van Vuuren, A. J., … Swart, H. C. (2019). Cu nanoclusters in ion exchanged soda-lime glass: Study of SPR and nonlinear optical behavior for photonics. Applied Materials Today, 15, 323–334. https://doi.org/10.1016/j.apmt.2019.02.016
Abràmoff, M. D., Magalhães, P. J., & Ram, S. J. (2004). Image processing with ImageJ. Biophotonics International, 11(7), 36–41. https://doi.org/10.1201/9781420005615.ax4
R Core Team. (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, 2(11), 1–3559.
RStudio: Integrated development environment for R. RStudio Team, Inc Boston, MA, USA: RStudio Team, & RStudio (2020). Team, Inc. Retrieved June 17, 2022, from https://www.rstudio.com/.
De Mendiburu, F. (2021). Agricolae: Statistical procedures for agricultural research. R Package Version 1.3-5. Retrieved June 27, 2022, from http://www.R-project.org.
Popat, R., & Banakara, K. (2020). Doebioresearch: Analysis of design of experiments for biological research. R package version 0.1.0. Retrieved July 4, 2021, from http://www.r-project.org/.
Achilonu, C. C., Gryzenhout, M., Ghosh, S., & Marais, G. J. (2023). In vitro evaluation of azoxystrobin, boscalid, fentin-hydroxide, propiconazole, pyraclostrobin fungicides against Alternaria alternata pathogen isolated from Carya illinoinensis in South Africa. Microorganisms, 11(7), 1691. https://doi.org/10.3390/MICROORGANISMS11071691.
Bongomin, F., Gago, S., Oladele, R. O., & Denning, D. W. (2017). Global and multi-national prevalence of fungal diseases - estimate precision. Journal of Fungi, 3(4). https://doi.org/10.3390/jof3040057
Ijadpanahsaravi, M., Teertstra, W. R., & Wösten, H. A. B. (2022). Inter- and intra-species heterogeneity in germination of Aspergillus conidia. Antonie Van Leeuwenhoek, 115(9), 1151–1164. https://doi.org/10.1007/s10482-022-01762-4
Pócsi, I., Giacometti, F., Ambrus, Á., & Logrieco, A. F. (2020). Editorial: Aspergillus-derived mycotoxins in the feed and food chain. Frontiers in Microbiology, 11. Retrieved from https://www.frontiersin.org/articles/https://doi.org/10.3389/fmicb.2020.606108.
Kanaujia, R., Singh, S., & Rudramurthy, S. M. (2023). Aspergillosis: An update on clinical spectrum, diagnostic schemes, and management. Current Fungal Infection Reports, 1–12. https://doi.org/10.1007/s12281-023-00461-5
Xu, S., Zhang, H., Qi, L., & Xiao, L. (2019). Conductometric acetone vapor sensor based on the use of gold-doped three-dimensional hierarchical porous zinc oxide microspheres. Mikrochimica Acta, 186(6), 342. https://doi.org/10.1007/s00604-019-3457-y
Kumar, P., Inwati, G. K., Mathpal, M. C., Ghosh, S., Roos, W. D., & Swart, H. C. (2021). Defects induced enhancement of antifungal activities of Zn doped CuO nanostructures. Applied Surface Science, 560(11), 150026-undefined. https://doi.org/10.1016/J.APSUSC.2021.150026
Kumar, P., Mathpal, M. C., Inwati, G. K., Kumar, S., Duvenhage, M. M., Roos, W. D., & Swart, H. C. (2023). Study of defect-induced chemical modifications in spinel zinc-ferrites nanostructures by in-depth XPS investigation. Magnetochemistry, 9(1), 20. https://doi.org/10.3390/magnetochemistry9010020
Kumar, P., Chandra Mathpal, M., Prakash, J., Viljoen, B. C., Roos, W. D., & Swart, H. C. (2020). Band gap tailoring of cauliflower-shaped CuO nanostructures by Zn doping for antibacterial applications. Journal of Alloys and Compounds, 832, 154968. https://doi.org/10.1016/j.jallcom.2020.154968
Prakash, J., Kumar, P., Harris, R. A., Swart, C., Neethling, J. H., van Vuuren, A. J., & Swart, H. C. (2016). Synthesis, characterization and multifunctional properties of plasmonic Ag-TiO2 nanocomposites. Nanotechnology, 27(35), 355707. https://doi.org/10.1088/0957-4484/27/35/355707
El-Batal, A. I., & Al Tamie, M. S. (2015). Biosynthesis of gold nanoparticles using marine Streptomyces cyaneus and their antimicrobial, antioxidant and antitumor (in vitro) activities. Journal of Chemical and Pharmaceutical Research, 7(7), 1020–1036.
Mishra, R. C., Kalra, R., Dilawari, R., Goel, M., & Barrow, C. J. (2022). Bio-synthesis of Aspergillus terreus mediated gold nanoparticle: Antimicrobial, antioxidant, antifungal and in vitro cytotoxicity studies. Materials, 15(11), 3877. https://doi.org/10.3390/ma15113877
Sheikh, H., & Awad, M. F. (2022). Biogenesis of nanoparticles with inhibitory effects on aflatoxin B1 production by Aspergillus flavus. Electronic Journal of Biotechnology, 60, 26–35. https://doi.org/10.1016/j.ejbt.2022.09.003
Jampílek, J., & Kráĺová, K. (2020). Chapter 14 - Impact of nanoparticles on toxigenic fungi. In M. Rai & K. A. Abd-Elsalam (Eds.), Nanomycotoxicology (pp. 309–348). Academic Press. https://doi.org/10.1016/B978-0-12-817998-7.00014-8.
Boroumand Moghaddam, A., Namvar, F., Moniri, M., Md. Tahir, P., Azizi, S., & Mohamad, R. (2015). Nanoparticles biosynthesized by fungi and yeast: A review of their preparation, properties, and medical applications. Molecules, 20(9), 16540–16565. https://doi.org/10.3390/molecules200916540
Khandel, P., & Shahi, S. K. (2018). Mycogenic nanoparticles and their bio-prospective applications: Current status and future challenges. Journal of Nanostructure in Chemistry, 8(4), 369–391. https://doi.org/10.1007/s40097-018-0285-2
Hendrickson, J. A., Hu, C., Aitken, S. L., & Beyda, N. (2019). Antifungal resistance: A concerning trend for the present and future. Current Infectious Disease Reports, 21(12), 1–8. https://doi.org/10.1007/S11908-019-0702-9/METRICS
Kareem, H. A., Samaka, H. M., & Abdulridha, W. M. (2021). Evaluation of the effect of the gold nanoparticles prepared by green chemistry on the treatment of cutaneous candidiasis. Current Medical Mycology, 7(1), 1–5. https://doi.org/10.18502/cmm.7.1.6176
Mikhailova, E. O. (2021). Gold nanoparticles: Biosynthesis and potential of biomedical application. Journal of Functional Biomaterials, 12(4), 70. https://doi.org/10.3390/jfb12040070
Manke, A., Wang, L., & Rojanasakul, Y. (2013). Mechanisms of nanoparticle-induced oxidative stress and toxicity. BioMed Research International, 2013, 942916. https://doi.org/10.1155/2013/942916.
Fuchs, J., Aghaei, M., Schachel, T. D., Sperling, M., Bogaerts, A., & Karst, U. (2018). Impact of the particle diameter on ion cloud formation from gold nanoparticles in ICPMS. Analytical Chemistry, 90(17), 10271–10278. https://doi.org/10.1021/acs.analchem.8b02007
Funding
Open access funding provided by University of the Free State.
Author information
Authors and Affiliations
Contributions
C.C.A. and P.K. conducted the experiments, performed data analysis, and wrote the main manuscript text. C.C.A. prepared Figs. 1, 8 and 9. P.K. prepared figures 2 - 7. H.C.S., W.D.R., and G.J.M. reviewed the manuscript and validated the data.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Informed Consent
None.
Competing Interests
The authors declare no competing interests.
Research Involving Humans and Animals Statement
None.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Achilonu, C.C., Kumar, P., Swart, H.C. et al. Zinc Oxide:Gold Nanoparticles (ZnO:Au NPs) Exhibited Antifungal Efficacy Against Aspergillus niger and Aspergillus candidus. BioNanoSci. 14, 799–813 (2024). https://doi.org/10.1007/s12668-024-01406-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12668-024-01406-z