Abstract
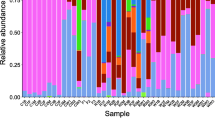
To investigate the differences in gut bacterial community of Parabramis pekinensis at different growth stages, we collected wild P. pekinensis from the Jingjiang region of the Yangtze River, and detected the intestinal microflora structure using high-throughput sequencing technology. Results show that during stage I the dominant bacteria were Proteobacteria, Actinobacteria, and Firmicutes. During stage II, the proportion of Proteobacteria and Actinobacteria decreased, while the proportion of Firmicutes and Fusobacteria increased, especially Clostridium and Cetobacteria increased significantly. During stage III, Cetobacterium had a dominant position, while the proportion of Firmicutes decreased slightly. In stage IV, the male and female fish showed obvious differences. In the female gut, the proportion of Proteobacteria increased to the first place, while Fusobacteria decreased to the second place. In the male fish, the proportion of Fusobacteria dropped to the fifth, especially that of Cetobacterium decreased significantly, and that of Verrucomicrobia increased. In stage V, the proportion of Fusobacteria increased again to the first place, while Proteobacteria did not decrease significantly in the female gut. The gut bacterial community in males changed into a structure similar to stage I. In stage VI, the gut bacterial community in both females and males changed into a structure similar to stage I. There were significant differences in the intestinal microflora structure of P. pekinensis at different gonad development stages and sexes. To some extent, the changes in intestinal microflora structure reflect the changes in the nutritional requirements of P. pekinensis.







Similar content being viewed by others
Availability of data and materials
The sequencing data were deposited in the NCBI Short Read Archive (SRA) with the accession number PRJNA706781.
References
Amnon A, Daniel M, Jose AN-M et al (2017) Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2(2):e00191-16
Austin B, Stuckey FL, Robertson PAW et al (1995) A probiotic strain of Vibrio alginolyticus effective in reducing diseases caused by Aeromonas salmonicida, Vibrio anguillarum and Vibrio ordalii. J Fish Dis 18(1):93–96
Bakke I, Skjermo J, Vo TA, Vadstein O (2013) Live feed is not a major determinant of the microbiota associated with cod larvae (Gadus morhua). Environ Microbiol Report 5(4):537–548
Carla PM, Fernando N-C, Paula S-M et al (2019) Sex, age, and bacteria: how the intestinal microbiota is modulated in a protandrous hermaphrodite fish. Front Microbiol 10:2512
Chong A, Ishak SD, Osman Z, Hashim R (2004) Effect of dietary protein level on the reproductive performance of female swordtails Xiphophorus helleri (Poeciliidae). Aquaculture 234(1–4):381–392
Christian Q, Elmar P, Pelin Y et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(D1):D590–D596
Das KM, Tripathi SD (1991) Studies on the digestive enzymes of grass carp, Ctenopharyngodon idella (Val.). Aquaculture 92:21–32
Eichmiller JJ, Hamilton MJ, Staley C et al (2016) Environment shapes the fecal microbiome of invasive carp species. Microbiome 4(1):44
Evan B, Ram RJ, Matthew RD et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37(8):852–857
Gregory CJ, Justin K, Jesse S et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Hailong G, Yaming F, Ya Z et al (2021) Differential study of the Parabramis pekinensis intestinal microbiota according to different habitats and different parts of the intestine. Annal Microbiol 71(1):1–11
Hjort HM, Hestbjerg HL, Jacob B et al (2014) High-resolution melt analysis for rapid comparison of bacterial community compositions. Appl Environ Microbiol 80(12):3568–3575
Huang X, Shi Z, Li W et al (2009) The aminoacids in different tissues of silver pomfret (Pampus argenteus) broodstock and their change with the gonad development. J Fish China 33(02):278–287 (in Chinese)
Jiang M, Xu M, Ying C et al (2020) The intestinal microbiota of lake anchovy varies according to sex, body size, and local habitat in Taihu lake. China Microbiologyopen 9(1):e00955
Joan C, Manuel C, Silvia Z et al (1994) Influence of nutritional composition of diet on sea bass, Dicentrarchus labrax L., reproductive performance and egg and larval quality. Aquaculture 128(3–4):345–361
Kashiwada K, Teshima S, Kanazawa A (1970) Studies on the production of B vitamins by intestinal bacteria of Fish-V evidence of the production of vitamin B12 by microorganisms in the intestinal canal of carp. Cyprinus Carpio Nippon Suisan Gakkaishi 36(4):421–424
Kessel MA, Dutilh BE, Neveling K et al (2011) Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.). AMB Express 1:41
Knight R, Vrbanac A, Taylor BC et al (2018) Best practices for analysing microbiomes. Nat Rev Microbiol 16(7):410–422
Larsen AM, Mohammed HH, Arias CR (2014) Characterization of the gut microbiota of three commercially valuable warmwater fish species. J Appl Microbiol 116(6):1396–1404
Lin Y, Jon A, Duane C et al (2014) Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J 8(3):541–551
Liu Y (1993) Reproduction physiology of Chinese cultured fish. Agriculture Press, Beijing (in Chinese)
Liu H, Guo X, Gooneratne R et al (2016) The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci Rep 6(1):24340
Lv D, Zhou Y, Ge Y et al (2018) Age structure and growth characteristics of Culter Ilishaeformis in Dianshan Lake. Acta Hydrobiol Sin 42(4):762–769 (in Chinese)
Nayak SK (2010) Role of gastrointestinal microbiota in fish. Aquac Res 41(11):1553–1573
Ni J, Yan Q, Yu Y, Zhang T (2014) Factors influencing the grass carp gut microbiome and its effect on metabolism. FEMS Microbiol Ecol 87:704–714
Nicola S, Jacques I, Levi W et al (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12(6):R60
Parma L, Candela M, Soverini M et al (2016) Next-generation sequencing characterization of the gut bacterial community of gilthead sea bream (Sparus aurata, L.) fed low fishmeal based diets with increasing soybean meal levels. Anim Feed Sci Technol 222:204–216
Parris DJ, Brooker RM, Morgan MA et al (2016) Whole gut microbiome composition of damselfish and cardinalfish before and after reef settlement. PeerJ 4:e2412
Pelin Y, Wegener PL, Pablo Y et al (2014) The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648
Rawls J, Samuel B, Gordon J (2004) Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci 101(13):4596–4601
Ringø E, Zhou Z, Vecino JG et al (2016) Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac Nutr 22(2):219–282
Romero J, Navarrete P (2006) 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of Coho Salmon (Oncorhynchus kisutch). Microb Ecol 51(4):422–430
Schloss PD (2021) Amplicon sequence variants artificially split bacterial genomes into separate clusters. mSphere 6(4):e0019121
Semova I, Carten JD et al (2012) Microbiota regulate intestinal absorption and metabolism of fatty acids in the Zebrafish. Cell Host Microbe 12(3):277–288
Sou M, Kamanda ND, Ulrich S (2015) Diet strongly influences the gut microbiota of surgeonfishes. Mol Ecol 24(3):656–672
Tandler A, Harel M, Koven WM, Kolkovski S (1995) Broodstock and larvae nutrition in gilthead seabream Sparus aurata-new findings on its mode of involvement in improving growth, survival and swimbladder inflation. Israeli J Aquac-Bamidgeh 47(3):95–111
Wang AR, Ran C, Ringø E, Zhou ZG (2018) Progress in fish gastrointestinal microbiota research. Rev Aquac 10(3):626–640
Ward NL, Steven B, Penn K et al (2009) Characterization of the intestinal microbiota of two Antarctic notothenioid fish species. Extremophiles 13(4):679–685
Watanabe T, Takeuchi T, Saito M, Nishimura K (1984) Effect of low protein-high calory of essential fatty acid deficiency diet on reproduction of rainbow trout. Nippon Suisan Gakkaishi 50(7):1207–1215
Wu S, Wang G, Angert ER et al (2012) Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS One 7:e30440
Yukgehnaish K, Kumar P, Sivachandran P et al (2020) Gut microbiota metagenomics in aquaculture: factors influencing gut microbiome and its physiological role in fish. Rev Aquac 12(3):1903–1927
Zarkasi KZ, Abell GCJ, Taylor RS et al (2014) Pyrosequencing-based characterization of gastrointestinal bacteria of Atlantic salmon (Salmo salar L.) within a commercial mariculture system. J Appl Microbiol 117(1):18–27
Acknowledgements
We would like to thank the members of the Taizhou Institute of Agricultural Sciences for their help. We also thank Novogene Co., Ltd. (Beijing, China) for providing sequencing services. We would like to thank Editage (www.editage.cn) for English language editing.
Funding
Jiangsu Agricultural Science and Technology Independent Innovation Fund “Study on the key technology of artificial larger-scale seedling breeding of Parabramis pekinensis” [No. CX(19)3011].
Author information
Authors and Affiliations
Contributions
HLG and YMF conceived and designed the study. HLG and ZJY collected the fish and prepared the samples. HLG drafted the manuscript YMF critically reviewed the manuscript and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
All procedures performed were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978) and approved by the Experimental Animal Ethics Committee of Taizhou Academy of Agricultural Sciences (Taizhou, China).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gu, H., Feng, Y. & Yang, Z. Differential study of the Parabramis pekinensis intestinal microbiota according to different gonad development stages. Fish Sci 88, 721–731 (2022). https://doi.org/10.1007/s12562-022-01631-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-022-01631-z