Abstract
Raw oysters are considered a culinary delicacy but are frequently the culprit in food-borne norovirus (NoV) infections. As commercial depuration procedures are currently unable to efficiently eliminate NoV from oysters, an optimisation of the process should be considered. This study addresses the ability of elevated water temperatures to enhance the elimination of NoV and Tulane virus (TuV) from Pacific oysters (Crassostrea gigas). Both viruses were experimentally bioaccumulated in oysters, which were thereafter depurated at 12 °C and 17 °C for 4 weeks. Infectious TuV and viral RNA were monitored weekly for 28 days by TCID50 and (PMAxx-) RT-qPCR, respectively. TuV RNA was more persistent than NoV and decreased by < 0.5 log10 after 14 days, while NoV reductions were already > 1.0 log10 at this time. For RT-qPCR there was no detectable benefit of elevated water temperatures or PMAxx for either virus (p > 0.05). TuV TCID50 decreased steadily, and reductions were significantly different between the two temperatures (p < 0.001). This was most evident on days 14 and 21 when reductions at 17 °C were 1.3–1.7 log10 higher than at 12 °C. After 3 weeks, reductions > 3.0 log10 were observed at 17 °C, while at 12 °C reductions did not exceed 1.9 log10. The length of depuration also had an influence on virus numbers. TuV reductions increased from < 1.0 log10 after seven days to > 4.0 log10 after 4 weeks. This implies that an extension of the depuration period to more than seven days, possibly in combination with elevated water temperatures, may be beneficial for the inactivation and removal of viral pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shellfish are commonly viewed as culinary delicacies, giving high retail prices. This is especially true for bivalve molluscs. Among those, raw oysters are the pinnacle of fine dining. Still, this high-class product is often associated with food-borne illness. In this context, norovirus (NoV) is of special relevance, as it is perceived as the most common cause of non-bacterial gastroenteritis globally. Even though NoV is known to spread directly from person to person, transmission through food is common. Shellfish, predominantly oysters, are repeatedly listed as the culprit in food-borne NoV infections (Bellou et al., 2013; Bitler et al., 2013; Guix et al., 2019; Hardstaff et al., 2018), and the occurrence of NoV outbreaks after the consumption of oyster dishes is well-documented (Baker et al., 2011; Iritani et al., 2014; Meghnath et al., 2019; Woods et al., 2016).
A widespread practice to remove pathogens is keeping oysters in tanks with clean sea water in a process termed depuration (Lee et al., 2008; McLeod et al., 2017b). This process has been shown to work well for the removal of bacterial pathogens. However, depuration is less effective for NoV, and outbreaks have repeatedly been linked to depurated oysters (McLeod et al., 2017a; Neish, 2013; Sharp et al., 2021; Shen et al., 2019). Allegedly, this is due to the binding of NoV particles to carbohydrate receptors present in the oyster digestive tissue (DT). These receptors closely resemble histo-blood group antigens (HBGAs) in the human intestine, to which NoV has been shown to bind (Huang et al., 2003; Le Guyader et al., 2006; Marionneau et al., 2002; Tian et al., 2006).
As commercially applied depuration is unable to efficiently prevent NoV infections associated with oyster consumption, modifications to the depuration process have been evaluated. The effects of light regimen, feeding, salinity and vibration have been examined. Also, depuration times have been extended for up to 6 weeks, and a promising approach may be elevated water temperatures (Battistini et al., 2021b; Drouaz et al., 2015; Kingsley et al., 2018; Martinez-Albores et al., 2020; Neish, 2013; Polo et al., 2018; Rupnik et al., 2021; Younger et al., 2020). A minimum of 8 °C is recommended for the depuration of Pacific oysters in the UK, but temperatures can vary depending on location. In China and parts of Australia water temperatures can even reach 25 °C (McLeod et al., 2017b). There is evidence that raising the water temperature increases the elimination of bacteria (Chae et al., 2009). As for virus removal, a benefit of elevated temperatures has also been suggested, but this may depend on the type of virus (Neish, 2013; Rupnik et al., 2021; Younger et al., 2020).
In the present study, a NoV GII.2 strain and its culturable surrogate Tulane virus (TuV) have been bioaccumulated in the Pacific oyster, Crassostrea gigas. This bivalve is invasive to Scandinavia and is considered both an intruder and a potential food resource. Contaminated oysters were placed in depuration tanks at two water temperatures to assess whether elevated temperatures improve the elimination of the two viruses. Infectious TuV as well as persistence of TuV and NoV RNA were monitored weekly over the course of 28 days.
Materials & Methods
Viruses & Cells
TuV strain M033 was provided by T. Farkas, Louisiana State University at Baton Rouge, LA, United States. LLC-MK2 cells (ATCC CCL-7) were grown in cell culture flasks with filter cap in Medium 199/Earle's salts/GlutaMAX (Thermo Scientific/Gibco, Waltham, MA, USA) with 10% foetal bovine serum (Thermo Scientific/Gibco) and 2% antibiotic/antimycotic solution (Thermo Scientific/Gibco) at 37 °C and 5% CO2. Cells were infected with TuV at a multiplicity of infection of 1 in M199 Earle's salts/GlutaMAX without additional supplements (maintenance medium). When a cytopathic effect was observed, TuV was harvested by 3× freeze-thawing. Debris was removed by centrifugation at 2500×g for 5 min. The supernatant was collected, and the titre of virus stock determined (3.1 × 107 TCID50/mL). The NoV GII.2 faecal sample was acquired from a child with acute gastroenteritis. The genotype was determined based on ORF2 sequencing and the Norovirus Automated Genotyping Tool (Kroneman et al., 2011).
Oysters
Oysters harvested near Nøtterøy, Vestfold and Telemark County, Norway, were provided by a local supplier. Oysters were size graded by the supplier and weighed 120 g on average.
Bioaccumulation and Depuration
Bioaccumulation was conducted in late June 2021 in natural sea water that was treated by UV irradiation. TuV and NoV stock was diluted 1:1000 in 60 L sea water to 3.1 × 104 TCID50/mL and 4.0 × 103 genome copies per millilitre (GC/mL), respectively. Two trays, each containing ~ 75 oysters were placed in tanks with sea water so that all were completely submerged. Bioaccumulation lasted for 20 h at a water temperature of ~ 12 °C and under constant aeration of the water. There was no feed added to the bioaccumulation tank. Oysters were rinsed in clean sea water and 15 oysters were collected to determine the initial virus concentration. The remaining oysters were divided equally and placed into two depuration tanks that differed in water temperature. Low temperature depuration was conducted at 12 ± 0.9 °C using incoming sea water routinely used in the depuration facility. Elevated temperature depuration was at 17 ± 0.2 °C. The water temperature was held stable by a swimming pool heat pump (Heat Splasher ECO Plug & Play 2,5 kW, model 1295). Depuration lasted for 4 weeks in a flow-through system without water recirculation.
Sample Processing and Virus Extraction
Samples were collected weekly until day 28. From each depuration tank at each sampling time, 15 oysters were collected, rinsed in clean sea water, and transported chilled to the laboratory within 3 h. Oysters were placed at 4 °C upon arrival for a maximum of 2 h until analysis. Oysters were shucked, DT removed and homogenised with scalpel blades. DT from five oysters was pooled to make one sample, resulting in three replicates per depuration temperature per sampling day. Virus was extracted from oyster tissue as described in Araud et al. (2016) with minor modifications. Homogenised tissue was aliquoted into 50 mL tubes and 5 mL PBS was added. The tissue was further homogenised with a pestle. Samples were centrifuged at 3000×g for 15 min. The supernatant was collected and immediately used for molecular and cell culture analysis. Excess virus extract was stored at − 80 °C.
50% Tissue Culture Infective Dose (TCID50)
LLC-MK2 cells were seeded into 96-well plates (~ 1 × 104 cells/well) and grown to confluency. Virus extracts were diluted ten-fold in maintenance medium. Fifty microlitre of these virus dilutions was inoculated into the wells in quadruplicate for incubation at 37 °C and 5% CO2 for 2 h. Maintenance medium (200 µL) was added, and plates were incubated at 37 °C and 5% CO2 until 7 days post infection. Cytopathic effect was converted to TCID50 per mL virus extract with the Spearman & Kärber algorithm described by Hierholzer and Killington (1996).
Propidium Monoazide (PMAxx) Treatment
PMAxx was included in the analysis to reduce PCR amplification of viruses with damaged capsids (Randazzo et al., 2018; Razafimahefa et al., 2021). PMAxx (20 mM, Biotium, Fremont, CA, USA) was diluted 1:10 in diH2O and added to 500 µL virus extract at a final concentration of 100 µM. Samples were incubated with shaking (150 rpm) at room temperature for 15 min in the dark. They were exposed to LED light (464–476 nm) for 15 min in the PhastBlue Photoactivation System for Tubes (GenIUL, Terrassa, Spain) at 100% intensity and placed on ice. Virus extracts that did not receive PMAxx treatment were kept on ice.
RNA-Extraction
RNA was isolated from 500 µL supernatant with NucliSens magnetic extraction reagents on a MiniMAG (BioMerieux, Marcy l'Etoile, France) as described in ISO 15216–1 (ISO, 2017). RNA was used for molecular analysis immediately after extraction or stored at − 80 °C.
One-Step RT-qPCR
The RNA UltraSense One-Step Quantitative RT-PCR System (Thermo Scientific/Applied Biosystems) was used at a reaction volume of 20 µL. TVIF primer and probe were chosen for TuV and COG2R/QNIF2d primers and QNISP probe for GII (Table 1). TVIF primer and probe concentrations were 0.4 and 0.05 µM, respectively. Concentrations of QNIF2d, COG2R and QNIFSP were 0.9, 0.5 and 0.25 µM. Cycling conditions were 55 °C/30 min, 95 °C/2 min, and 45 cycles of 95 °C/15 s, 55 °C/20 s and 64 °C/40 s for TuV. NoV was run at 55 °C/30 min, 95 °C/2 min, and 45 cycles of 95 °C/15 s, 60 °C/60 s, 64 °C/8 s and 68 °C/8 s.
Droplet Digital RT-PCR (RT-ddPCR)
For quantification of genome copies (GC) per mL after RT-qPCR, NoV and TuV positive controls were quantified in RT-ddPCR. The One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad, Hercules, CA, USA) and the QX200 Droplet Digital System (Bio-Rad) were used. The NoV stock was determined to be 4.0 × 106 GC/mL according to the method described by Persson et al. (2018). QNIFSP and TVIF probes were modified with a ZEN/Iowa Black FQ quencher (Table 1). Primer and probe concentrations were adjusted to 0.9 and 0.25 µM, respectively. TuV cycling conditions were 48 °C/60 min, 95 °C/10 min, and 39 cycles of 95 °C/30 s and 55 °C/60 s with a ramp speed of 2 °C/s, and a final elongation step of 98 °C/10 min. For NoV, annealing temperature was reduced to 53 °C and ramp speed was 3 °C/s. The QuantaSoft™ software (Bio-Rad), was used for data analysis and the accepted number of droplets generated was ≥ 10,000.
Data Analysis
The experiment was conducted once in summer 2021. Three biological replicates were analysed at each sampling point. RT-qPCR was run on technical duplicates. Relative viral RNA copy numbers were estimated using standard curves from ten-fold dilution series of stock virus RNA. Copy numbers were expressed with the following formula: \({N}_{1}={N}_{2}\times {\left(1+E\right)}^{\left({Ct}_{2}-{Ct}_{1}\right)}\). N1 and N2 are the amount of viral RNA in the sample and in the positive control, respectively; Ct1 and Ct2 are threshold cycles for sample and control, respectively; E is the efficiency of amplification. These numbers were the basis for calculating log10 reductions as follows: log10 reduction = log10 (control virus) – log10 (inactivated virus). ANOVA and Tukey HSD post-hoc test was conducted in RStudio version 1.3.959 (R Core Team, 2018; RStudio Team, 2020). P values < 0.05 were deemed significant.
Results
Persistence of Viral RNA (TuV, NoV)
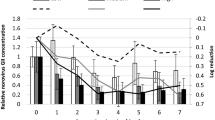
RT-qPCR underestimated the reduction in infectious TuV compared to TCID50 (Fig. 1a, b). TuV RNA decreased by < 1.0 log10 after 28 days. After 3 weeks, a TuV reduction of ~ 0.3 log10 was observed at low water temperature. At the higher water temperature, a 0.3 log10 reduction in TuV RNA occurred one week earlier. Still, no significant difference in reduction could be detected between the two temperatures (p > 0.05).
Concentration of TuV (solid line) and NoV (dashed line) RNA in genome copies per mL as determined by RT-qPCR without the addition of PMAxx (a), as well as TuV titre in TCID50 per mL (b) during the depuration period at high (filled circle) and low (empty diamond) water temperatures; the ribbon represents standard error of three replicates, * (asterisk) = Below the limit of detection (LOD)
NoV GII decreased by 0.8 log10 after week one (Fig. 1a). Prolonged depuration (≥ 14 days) resulted in a ≥ 1.0 log10 reduction in NoV. Over the course of the depuration period, an average reduction of 1.3 log10 was observed, but there was no significant difference in reduction between the two temperatures (p = 0.057).
There was no observable benefit of applying PMAxx for differentiating between infectious and non-infectious virus for either TuV or NoV (p > 0.05, data not shown). PMAxx data is available as a supplementary file online.
Reduction in Infectious Virus (TuV)
TuV titre was monitored throughout the 4-week depuration period. The initial TuV concentration was 3.0 × 104 TCID50 per mL supernatant (Fig. 1b). Regardless of water temperature, TuV decreased by < 0.8 log10 within the 1st week to 6.7 × 103 and 1.5 × 104 TCID50/mL at high and low temperatures, respectively. Towards the end of the experiment, a reduction of ~ 4.2 log10 to < 10 TCID50/mL was recorded at 12 °C, while at 17 °C TuV was reduced by 4.5 log10 to below the limit of detection. Overall, there was a significant difference in reduction of infectious TuV at high and low temperature (p < 0.001). This was most evident on days 14 and 21 (p < 0.001, p = 0.005).
Discussion
Depuration has shown a limited ability to remove NoV from oysters. One important factor influencing virus removal rates may be water temperature during depuration. Lower temperatures lead to low metabolic activity in oysters and seem to decrease virus removal rates (Lees et al., 2010). Therefore, the elimination of infectious viruses may increase at higher water temperatures.
Our data suggest a significant effect of the elevated water temperature on infectious TuV from week 2 onwards. TCID50 reductions on day 14 were 2.6 and 1.0 log10 at 17 °C and 12 °C, respectively. After 3 weeks, reductions > 3.0 log10 were observed at the higher water temperature, while at the lower temperature reductions were still < 2.0 log10. After 4 weeks, no infectious TuV was detected at 17 °C, but small numbers of infectious TuV remained at the low temperature. The benefit of elevated temperatures during depuration for the reduction in virus infectivity has been illustrated previously (Kingsley et al., 2018; Neish, 2013).
Contrary to TuV infectivity, we found no advantage of the higher temperature for the reduction in either TuV or NoV RNA. Correspondingly, Polo et al. (2018) report that TuV RNA levels in Pacific oysters were comparable at temperatures between 9 and 17 °C. Still, for the reduction in NoV RNA, a beneficial effect of elevated temperatures has been suggested (Choi & Kingsley, 2016; Neish, 2013; Rupnik et al., 2021; Younger et al., 2020). It is conceivable that a greater difference in water temperature results in a greater difference in depuration kinetics (Rupnik et al., 2021). Our experiment was conducted in June to July, so the sea water temperature was already ≥ 11.5 °C. The ambient water temperature was kept as is after entering the depuration facility, a common practice for the supplier, and was directly used for the low temperature tank. To get an impression of the effect of elevated water temperatures during colder months, additional data should be gathered in winter to complement present results. During this time of year NoV is more prevalent in oysters (EFSA, 2019) and sea water temperatures can fall below 5 °C in Scandinavian waters.
Not only did the elevated water temperature have no effect on the removal of virus RNA, but RNA levels remained comparatively stable throughout the 4-week trial. Likewise, virus RNA decreased within the 1st days and no benefit of prolonged depuration for the reduction in Caliciviridae RNA was observed in related work (Battistini et al., 2021b; Choi & Kingsley, 2016; Rupnik et al., 2021; Ueki et al., 2007). Similarly, we observed the highest reductions in NoV RNA within the 1st week. For the remainder of the experiment, NoV levels were rather stable. A decrease in RNA during the 1st week and no noticeable reduction afterwards may reflect that unbound NoV was washed out of the oysters briefly after bioaccumulation, whereas bound virus remained within oysters for the rest of the study. Conversely, a continuous decline in TuV and NoV GI RNA could be demonstrated by Drouaz et al. (2015) and Polo et al. (2018). The stability of TuV RNA during our depuration trial, contrary to the steady decrease in infectious TuV, may indicate virus inactivation in the oyster without significant virus elimination, as previously suggested by Leduc et al. (2020). In contrast, Polo et al. (2018) detected similar reductions in TuV infectivity and RNA, which implies TuV elimination. These discrepancy underlines the importance of consulting data on virus infectivity in addition to RT-qPCR data. To come to a consensus on the fate of TuV in oysters, further studies are needed.
Just like there was no advantage of the elevated water temperature on RNA removal, we did not find any effect of PMAxx for either virus. However, application of the dye has been advantageous over regular RT-qPCR in distinguishing infectious from non-infectious NoV in oysters (Randazzo et al., 2018). Randazzo et al. (2018) examined the effect of PMAxx on heat treated virus, and they extracted NoV according to the ISO standard, which includes incubation at 37 °C and 60 °C (ISO, 2017). These temperatures would have facilitated the entry of PMAxx due to induced capsid damage. During depuration in the present study the capsid of TuV and NoV was most likely not exposed to conditions that induce severe capsid damage. Accordingly, infectious TuV declined in TCID50, whereas no effect on the RNA or of PMAxx was observed.
In the present work we also observed a more rapid decline in NoV compared to TuV RNA during the 1st week. NoV RNA was already reduced by ~ 0.8 log10 after the 1st week, whereas TuV RNA was reduced by < 0.8 log10 at the end of the experimental period. Several factors may contribute to this, among them the conditions encountered during bioaccumulation or a difference in the efficiency of virus recovery from oyster tissue. We performed the bioaccumulation of both viruses simultaneously, so there may have been competition over HBGA-binding. TuV may have outcompeted NoV in binding since TuV concentration during bioaccumulation exceeded that of NoV. Additionally, TuV may have a greater binding affinity than NoV GII.2. As a result, the majority of NoV may have been present as an unbound fraction in the oyster DT and would have been eliminated quickly. In oysters, Ueki et al. (2021) report unsuccessful bioaccumulation of GII.2, whereas Langlet et al. (2015) confirmed at least a low binding capacity to homogenised oyster tissue. Moreover, seasonal variations in NoV binding to oyster tissue were demonstrated by Maalouf et al. (2010). They suggest that HBGA expression is higher during late winter and spring. As mentioned, the present study was conducted in mid-summer, so that HBGA expression in oyster DT may have been decreased and might have influenced virus binding. Nevertheless, NoV must have bound to receptors, as NoV RNA was detected throughout the experiment. A possible difference in binding affinity could be further investigated and a different NoV strain could be chosen for future work.
The reductions in NoV RNA observed in the present study may result in a safe product, but this will depend on the initial level of NoV contamination. According to the European Food Safety Authority (EFSA) NoV contamination levels in oysters averaged at 337 GC/g, and only 5.5% of samples were > 1000 GC/g (EFSA, 2019). In the present depuration trial, ~ 0.8 and 1.0 log10 of the initial NoV load was removed after 1 and 2 weeks, respectively. According to these reductions and the EFSA data, NoV could be reduced to ≤ 100 GC/g during a 2 week depuration. Such a low concentration in oyster DT has not been involved in NoV outbreaks according to Lowther et al. (2012). If we assume that the reduction in NoV infectivity follows a similar pattern as TuV, a 1.0 to 2.6 log10 decrease in infectious NoV might be expected after 14 days, depending on water temperature. Such reductions would reduce the risk of NoV infections, if NoV contamination levels are as low as the average value stated in EFSA (2019). While these numbers provide an estimate to work with, the depuration rates may vary with NoV levels and with the amount of time that has passed since contamination. In case of recent contamination most NoV may be unbound and therefore easier and faster to remove, whereas NoV that was present in the tissue for a longer time may be bound to HBGAs and thus more difficult to eliminate. In addition, EFSA (2019) determined that 1.2% of analysed samples contained > 5000 NoV GC/g, which may be encountered after recent contamination or during winter (Battistini et al., 2021a). In that case, the reductions observed in the present study may not suffice to ensure a safe product, and depuration may have to be extended to 1 month, as we observed a > 4.0 log10 reduction in infectious TuV after this time. The reductions in infectious TuV and NoV in oysters should be further examined, for instance by utilising the human intestinal enteroid system (Ettayebi et al., 2016) or human volunteer studies (Richards, 2012), to make sure the two viruses are comparable.
Overall, we observed an advantage of elevated water temperatures for the reduction in infectious TuV. This was noticeable after 14 days when TuV was reduced by 1.0 log10 and 2.6 log10 at the low and elevated temperature, respectively. After 7 days, reductions were not noticeably different. This implies elevated water temperatures may primarily be relevant for a 2- to 3-week depuration. After 28 days, we detected reductions in TuV infectivity of > 4.0 log10, regardless of temperature. In case of suspected recent faecal contamination and high virus concentrations, an extended depuration of 3 to 4 weeks could be considered. The economic burden of prolonging the depuration period must be weighed against the burden of heating up the depuration water.
Data Availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Araud, E., DiCaprio, E., Ma, Y., Lou, F., Gao, Y., Kingsley, D., Hughes, J. H., & Li, J. (2016). Thermal inactivation of enteric viruses and bioaccumulation of enteric foodborne viruses in live oysters (Crassostrea virginica). Applied and Environmental Microbiology, 82(7), 2086–2099. https://doi.org/10.1128/aem.03573-15
Baker, K., Morris, J., McCarthy, N., Saldana, L., Lowther, J., Collinson, A., & Young, M. (2011). An outbreak of norovirus infection linked to oyster consumption at a UK restaurant, february 2010. Journal of Public Health, 33(2), 205–211. https://doi.org/10.1093/pubmed/fdq089
Battistini, R., Listorti, V., Squadrone, S., Pederiva, S., Abete, M. C., Mua, R., Ciccotelli, V., Suffredini, E., Maurella, C., Baioni, E., Orlandi, M., Ercolini, C., & Serracca, L. (2021a). Occurrence and persistence of enteric viruses, arsenic and biotoxins in pacific oysters farmed in an italian production site. Marine Pollution Bulletin. https://doi.org/10.1016/j.marpolbul.2020.111843
Battistini, R., Masotti, C., Listorti, V., Suffredini, E., Maurella, C., Garcia-Vozmediano, A., Costa, E., Iacona, F., Orlandi, M., Ercolini, C., & Serracca, L. (2021b). Norovirus persistence in oysters to prolonged commercial purification. Pathogens. https://doi.org/10.3390/pathogens10080944
Bellou, M., Kokkinos, P., & Vantarakis, A. (2013). Shellfish-borne viral outbreaks: A systematic review. Food and Environmental Virology, 5(1), 13–23. https://doi.org/10.1007/s12560-012-9097-6
Bitler, E. J., Matthews, J. E., Dickey, B. W., Eisenberg, J. N., & Leon, J. S. (2013). Norovirus outbreaks: A systematic review of commonly implicated transmission routes and vehicles. Epidemiology and Infection, 141(8), 1563–1571. https://doi.org/10.1017/s095026881300006x
Chae, M. J., Cheney, D., & Su, Y. C. (2009). Temperature effects on the depuration of Vibrio parahaemolyticus and Vibrio vulnificus from the American Oyster (Crassostrea virginica). Journal of Food Science, 74(2), 62–66. https://doi.org/10.1111/j.1750-3841.2008.01031.x
Choi, C., & Kingsley, D. H. (2016). Temperature-dependent persistence of human norovirus within oysters (Crassostrea virginica). Food and Environmental Virology, 8(2), 141–147. https://doi.org/10.1007/s12560-016-9234-8
Drouaz, N., Schaeffer, J., Farkas, T., Le Pendu, J., & Le Guyader, F. S. (2015). Tulane virus as a potential surrogate to mimic norovirus behavior in oysters. Applied and Environmental Microbiology, 81(15), 525249–525256. https://doi.org/10.1128/AEM.01067-15
EFSA. (2019). Analysis of the European Baseline survey of norovirus in oysters. EFSA Journal. https://doi.org/10.2903/j.efsa.2019.5762
Ettayebi, K., Crawford, S. E., Murakami, K., Broughman, J. R., Karandikar, U., Tenge, V. R., Neill, F. H., Blutt, S. E., Zeng, X. L., Qu, L., Kou, B., Opekun, A. R., Burrin, D., Graham, D. Y., Ramani, S., Atmar, R. L., & Estes, M. K. (2016). Replication of human noroviruses in stem cell-derived human enteroids. Science, 353(6306), 1387–1393. https://doi.org/10.1126/science.aaf5211
Guix, S., Pintó, R., & Bosch, A. (2019). Final consumer options to control and prevent foodborne norovirus infections. Viruses. https://doi.org/10.3390/v11040333
Hardstaff, J. L., Clough, H. E., Lutje, V., McIntyre, K. M., Harris, J. P., Garner, P., & O’Brien, S. J. (2018). Foodborne and food-handler norovirus outbreaks: A systematic review. Foodborne Pathogens and Disease, 15(10), 589–597. https://doi.org/10.1089/fpd.2018.2452
Hierholzer, J. C., & Killington, R. A. (1996) Virus Isolation and Quantitation. In B. W. J. Mahy, & H. O. Kangro (Eds.), Virology Methods Manual (1st ed., pp. 25-46). Academic Press Limited
Huang, P., Farkas, T., Marionneau, S., Zhong, W., Ruvoen-Clouet, N., Morrow, A. L., Altaye, M., Pickering, L. K., Newburg, D. S., LePendu, J., & Jiang, X. (2003). Noroviruses bind to human ABO, lewis, and secretor histo-blood group antigens: Identification of 4 distinct strain-specific patterns. Journal of Infectious Diseases, 188, 19–31. https://doi.org/10.1086/375742
Iritani, N., Kaida, A., Abe, N., Kubo, H., Sekiguchi, J., Yamamoto, S. P., Goto, K., Tanaka, T., & Noda, M. (2014). Detection and genetic characterization of human enteric viruses in oyster-associated gastroenteritis outbreaks between 2001 and 2012 in Osaka City. Japan. Journal of Medical Virology, 86(12), 2019–2025. https://doi.org/10.1002/jmv.23883
ISO (2017). ISO 15216–1:2017 Microbiology of the Food Chain - Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR - Part 1: Method for Quantification (15216–1). Geneva, Switzerland: International Organization for Standardization (ISO). https://doi.org/10.31030/2600632
Kageyama, T., Kojima, S., Shinohara, M., Uchida, K., Fukushi, S., Hoshino, F. B., Takeda, N., & Katayama, K. (2003). Broadly reactive and highly sensitive assay for norwalk-like viruses based on real-time quantitative reverse transcription-PCR. Journal of Clinical Microbiology, 41(4), 1548–1557. https://doi.org/10.1128/jcm.41.4.1548-1557.2003
Kingsley, D. H., Chen, H., & Meade, G. K. (2018). Persistence of MS-2 bacteriophage within Eastern Oysters. Food and Environmental Virology, 10(1), 83–88. https://doi.org/10.1007/s12560-017-9315-3
Kroneman, A., Vennema, H., Deforche, K. V. D., Avoort, H. V. D., Peñaranda, S., Oberste, M. S., Vinjé, J., & Koopmans, M. (2011). An Automated genotyping tool for enteroviruses and noroviruses. Journal of Clinical Virology, 51(2), 121–125. https://doi.org/10.1016/j.jcv.2011.03.006
Langlet, J., Kaas, L., & Greening, G. (2015). Binding-based RT-qPCR assay to assess binding patterns of noroviruses to shellfish. Food and Environmental Virology. https://doi.org/10.1007/s12560-015-9180-x
Le Guyader, F. S., Loisy, F., Atmar, R. L., Hutson, A. M., Estes, M. K., Ruvoën-Clouet, N., Pommepuy, M., & Le Pendu, J. (2006). Norwalk virus-specific binding to oyster digestive tissues. Emerging Infectious Diseases, 12(6), 931–936. https://doi.org/10.3201/eid1206.051519
Leduc, A., Leclerc, M., Challant, J., Loutreul, J., Robin, M., Maul, A., Majou, D., Boudaud, N., & Gantzer, C. (2020). F-Specific RNA bacteriophages model the behavior of human noroviruses during purification of oysters: The main mechanism is probably inactivation rather than release. Applied and Environmental Microbiology. https://doi.org/10.1128/aem.00526-20
Lee, R., Lovatelli, A., & Ababouch, L. (2008). Bivalve Depuration: Fundamental and Practical Aspects. FAO Fisheries Technical Paper (511, pp. 41–64). Rome, Italy: FAO.
Lees, D., Younger, A., & Doré, B. (2010). Depuration and Relaying. In G. Rees, K. Pond, D. Kay, J. Bartram, & J. Santo Domingo (Eds.), Safe Management of Shellfish and Harvest Waters: Minimizing Health Risks from Sewage-Contaminated Shellfish (1st ed., pp. 145–181). IWA Publishing on behalf of WHO
Loisy, F., Atmar, R. L., Guillon, P., Le Cann, P., Pommepuy, M., & Le Guyader, F. S. (2005). Real-time RT-PCR for norovirus screening in shellfish. Journal of Virological Methods, 123(1), 1–7. https://doi.org/10.1016/j.jviromet.2004.08.023
Lowther, J. A., Gustar, N. E., Hartnell, R. E., & Lees, D. N. (2012). Comparison of norovirus RNA levels in outbreak-related oysters with background environmental levels. Journal of Food Protection, 75(2), 389–393. https://doi.org/10.4315/0362-028X.JFP-11-360
Maalouf, H., Zakhour, M., Le Pendu, J., Le Saux, J. C., Atmar, R. L., & Le Guyader, F. S. (2010). Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters. Applied and Environmental Microbiology, 76(16), 5621–5630. https://doi.org/10.1128/aem.00148-10
Marionneau, S., Ruvoën, N., Le Moullac-Vaidye, B., Clement, M., Cailleau-Thomas, A., Ruiz-Palacois, G., Huang, P., Jiang, X., & Le Pendu, J. (2002). Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology, 122(7), 1967–1977. https://doi.org/10.1053/gast.2002.33661
Martinez-Albores, A., Lopez-Santamarina, A., Rodriguez, J. A., Ibarra, I. S., Mondragón, A. D. C., Miranda, J. M., Lamas, A., & Cepeda, A. (2020). Complementary methods to improve the depuration of bivalves: A review. Foods. https://doi.org/10.3390/foods9020129
McLeod, C., Polo, D., Le Saux, J.-C., & Le Guyader, F. S. (2017b). Final Report: Evaluating the Effectiveness of Depuration in Removing Norovirus from Oysters (FS101068). UK: Seafood Safety Assessment Limited and Ifremer - Commissioned by the UK Food Standards Agency.
McLeod, C., Polo, D., Le Saux, J.-C., & Le Guyader, F. S. (2017a). Depuration and relaying: A review on potential removal of norovirus from oysters. Comprehensive Reviews in Food Science and Food Safety, 16(4), 692–706. https://doi.org/10.1111/1541-4337.12271
Meghnath, K., Hasselback, P., McCormick, R., Prystajecky, N., Taylor, M., McIntyre, L., Man, S., Whitfield, Y., Warshawsky, B., McKinley, M., Bitzikos, O., Hexemer, A., & Galanis, E. (2019). Outbreaks of norovirus and acute gastroenteritis associated with British Columbia Oysters, 2016–2017. Food and Environmental Virology, 11(2), 138–148. https://doi.org/10.1007/s12560-019-09374-4
Neish, A. (2013). Investigative Trials on the Purification of Oysters to Identify Ways of Reducing Norovirus. Cefas contract report C5224. Dorset, UK: Centre for Environment, Fisheries & Aquaculture Science (Cefas).
Persson, S., Eriksson, R., Lowther, J., Ellström, P., & Simonsson, M. (2018). Comparison between RT droplet digital PCR and RT Real-Time PCR for quantification of noroviruses in oysters. International Journal of Food Microbiology, 284, 73–83. https://doi.org/10.1016/j.ijfoodmicro.2018.06.022
Polo, D., Schaeffer, J., Teunis, P., Buchet, V., & Le Guyader, F. S. (2018). Infectivity and RNA persistence of a norovirus surrogate, the tulane virus, in oysters. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2018.00716
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/
Randazzo, W., Khezri, M., Ollivier, J., Le Guyader, F. S., Rodríguez-Díaz, J., Aznar, R., & Sánchez, G. (2018). Optimization of PMAxx pretreatment to distinguish between human norovirus with intact and altered capsids in shellfish and sewage samples. International Journal of Food Microbiology. https://doi.org/10.1016/j.ijfoodmicro.2017.11.011
Razafimahefa, R. M., Ludwig-Begall, L. F., Le Guyader, F. S., Farnir, F., Mauroy, A., & Thiry, E. (2021). Optimisation of a PMAxx-RT-qPCR assay and the preceding extraction method to selectively detect infectious murine norovirus particles in mussels. Food and Environmental Virology. https://doi.org/10.1007/s12560-020-09454-w
Richards, G. P. (2012). Critical review of norovirus surrogates in food safety research: Rationale for considering volunteer studies. Food and Environmental Virology, 4(1), 6–13. https://doi.org/10.1007/s12560-011-9072-7
RStudio Team (2020). RStudio: Integrated Development Environment for R. Boston, MA, USA: RStudio PBC. https://www.rstudio.com/
Rupnik, A., Dore, W., Devilly, L., Fahy, J., Fitzpatrick, A., Schmidt, W., Hunt, K., Butler, F., & Keaveney, S. (2021). Evaluation of norovirus reduction in environmentally contaminated pacific oysters during laboratory controlled and commercial depuration. Food and Environmental Virology, 13, 229–240. https://doi.org/10.1007/s12560-021-09464-2
Sharp, J. H., Clements, K., Diggens, M., McDonald, J. E., Malham, S. K., & Jones, D. L. (2021). E. coli is a poor end-product criterion for assessing the general microbial risk posed from consuming norovirus contaminated shellfish. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2021.608888
Shen, X., Hou, Y., Su, Y. C., Liu, C., Oscar, T., & DePaola, A. (2019). Efficacy of Vibrio parahaemolyticus depuration in oysters (Crassostrea gigas). Food Microbiology, 79, 35–40. https://doi.org/10.1016/j.fm.2018.10.005
Tian, P., Bates, A. H., Jensen, H. M., & Mandrell, R. E. (2006). Norovirus binds to blood group A-like antigens in oyster gastrointestinal cells. Letters in Applied Microbiology, 43(6), 645–651. https://doi.org/10.1111/j.1472-765x.2006.02010.x
Ueki, Y., Amarasiri, M., Kamio, S., Sakagami, A., Ito, H., Uprety, S., Umam, A. N., Miura, T., Nguyen, T. H., & Sano, D. (2021). Human norovirus disease burden of consuming Crassostrea gigas oysters: A case-study from Japan. Food Control. https://doi.org/10.1016/j.foodcont.2020.107556
Ueki, Y., Shoji, M., Suto, A., Tanabe, T., Okimura, Y., Kikuchi, Y., Saito, N., Sano, D., & Omura, T. (2007). Persistence of caliciviruses in artificially contaminated oysters during depuration. Applied and Environmental Microbiology, 73(17), 5698–5701. https://doi.org/10.1128/aem.00290-07
Woods, J. W., Calci, K. R., Marchant-Tambone, J. G., & Burkhardt, W., 3rd. (2016). Detection and molecular characterization of norovirus from oysters implicated in outbreaks in the US. Food Microbiology, 59, 76–84. https://doi.org/10.1016/j.fm.2016.05.009
Younger, A. D., Neish, A., Walker, D. I., Jenkins, K. L., Lowther, J. A., Stapleton, T. A., & Alves, M. T. (2020). Strategies to reduce norovirus (NoV) contamination from oysters under depuration conditions. Food and Chemical Toxicology. https://doi.org/10.1016/j.fct.2020.111509
Funding
Open Access funding provided by Institute Of Marine Research. The research covered in this publication were financed by the Institute of Marine Research, Bergen, Norway, and partial financial support was received from Norwegian Shores AS, Nøtterøy, Norway. Gratitude goes to Norwegian Shores AS for the supply and transport of Pacific oysters and for providing facilities for this study.
Author information
Authors and Affiliations
Contributions
SMS: Conceptualization, data curation, formal analysis, investigation, methodology, validation, visualisation, writing original draft, writing review and editing; AD: Conceptualization, funding acquisition, methodology, project administration, supervision, validation, writing review and editing; MK: Data curation, investigation, validation; BTL: Conceptualization, funding acquisition, methodology, project administration, supervision, validation, writing review and editing; MM: Conceptualization, formal analysis, funding acquisition, investigation, methodology, project administration, supervision, validation, writing review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stoppel, S.M., Duinker, A., Khatri, M. et al. Temperature Dependent Depuration of Norovirus GII and Tulane Virus from Oysters (Crassostrea gigas). Food Environ Virol 15, 43–50 (2023). https://doi.org/10.1007/s12560-022-09547-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-022-09547-8