Abstract
Gene regulatory network (GRN) inference based on single-cell RNA sequencing data (scRNAseq) plays a crucial role in understanding the regulatory mechanisms between genes. Various computational methods have been employed for GRN inference, but their performance in terms of network accuracy and model generalization is not satisfactory, and their poor performance is caused by high-dimensional data and network sparsity. In this paper, we propose a self-supervised method for gene regulatory network inference using single-cell RNA sequencing data (CVGAE). CVGAE uses graph neural network for inductive representation learning, which merges gene expression data and observed topology into a low-dimensional vector space. The well-trained vectors will be used to calculate mathematical distance of each gene, and further predict interactions between genes. In overall framework, FastICA is implemented to relief computational complexity caused by high dimensional data, and CVGAE adopts multi-stacked GraphSAGE layers as an encoder and an improved decoder to overcome network sparsity. CVGAE is evaluated on several single cell datasets containing four related ground-truth networks, and the result shows that CVGAE achieve better performance than comparative methods. To validate learning and generalization capabilities, CVGAE is applied in few-shot environment by change the ratio of train set and test set. In condition of few-shot, CVGAE obtains comparable or superior performance.
Graphical Abstract
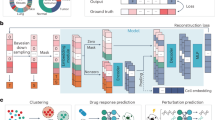
CVGAE utilizes a beta variational autoencoder framework in conjunction with graph neural networks to characterize the underlying gene regulatory networks in single-cell gene expression data. The model employs multiple stacked SAGE layers to produce embedding representations of domain nodes, ensuring that the vector representation adheres to a multivariate Gaussian distribution. CVGAE leverages further convolutional computation and multi-layer perceptrons to determine the strength of interactions between nodes.









Similar content being viewed by others
References
Haghverdi L, Büttner M, Wolf FA et al (2016) Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods 13(10):845–848. https://doi.org/10.1038/nmeth.3971
Muzio G, O’Bray L, Borgwardt K (2021) Biological network analysis with deep learning. Brief Bioinform 22(2):1515–1530. https://doi.org/10.1093/bib/bbaa257
Liu W, Sun X, Yang L et al (2022) NSCGRN: a network structure control method for gene regulatory network inference. Brief Bioinform 23(5):bbac156. https://doi.org/10.1093/bib/bbac156
Liu W, Yang Y, Lu X et al (2023) NSRGRN: a network structure refinement method for gene regulatory network inference. Brief Bioinform 24(3):bbad129. https://doi.org/10.1093/bib/bbad129
Nguyen H, Tran D, Tran B et al (2021) A comprehensive survey of regulatory network inference methods using single cell RNA sequencing data. Brief Bioinform 22(3):bbaa190. https://doi.org/10.1093/bib/bbaa190
Liu W, Jiang Y, Peng L et al (2022) Inferring gene regulatory networks using the improved markov blanket discovery algorithm. Interdiscip Sci 14(1):168–181. https://doi.org/10.1007/s12539-021-00478-9
Woodhouse S, Piterman N, Wintersteiger CM et al (2018) SCNS: a graphical tool for reconstructing executable regulatory networks from single-cell genomic data. BMC Syst Biol 12(1):59–59. https://doi.org/10.1186/s12918-018-0581-y
Matsumoto H, Kiryu H, Furusawa C et al (2017) SCODE: an efficient regulatory network inference algorithm from single-cell RNA-Seq during differentiation. Bioinformatics 33(15):2314–2321. https://doi.org/10.1093/bioinformatics/btx194
Matsumoto H, Kiryu H (2016) SCOUP: a probabilistic model based on the Ornstein-Uhlenbeck process to analyze single-cell expression data during differentiation. BMC Bioinform 17(1):232–232. https://doi.org/10.1186/s12859-016-1109-3
Liu H, Li P, Zhu M et al (2016) Nonlinear network reconstruction from gene expression data using marginal dependencies measured by DCOL. PLoS ONE 11(7):e0158247–e0158247. https://doi.org/10.1371/journal.pone.0158247
Aibar S, González-Blas CB, Moerman T et al (2017) SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14(11):1083–1086. https://doi.org/10.1038/nmeth.4463
Huynh-Thu VA, Irrthum A, Wehenkel L et al (2010) Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5(9):e12776–e12776. https://doi.org/10.1371/journal.pone.0012776
Shu H, Zhou J, Lian Q et al (2021) Modeling gene regulatory networks using neural network architectures. Nat Comput Sci 1(7):491–501. https://doi.org/10.1038/s43588-021-00099-8
Yuan Y, Bar-Joseph Z (2021) Deep learning of gene relationships from single cell time-course expression data. Brief Bioinform 22(5):bbab142. https://doi.org/10.1093/bib/bbab142
Kc K, Li R, Cui F et al (2019) GNE: a deep learning framework for gene network inference by aggregating biological information. BMC Syst Biol 13(2):1–14. https://doi.org/10.1093/bioinformatics/btac559
Yue X, Wang Z, Huang J et al (2020) Graph embedding on biomedical networks: methods, applications and evaluations. Bioinformatics 36(4):1241–1251. https://doi.org/10.1093/bioinformatics/btz718
Zhao M, He W, Tang J et al (2022) A hybrid deep learning framework for gene regulatory network inference from single-cell transcriptomic data. Brief Bioinform 23(2):b568–b568. https://doi.org/10.1093/bib/bbab568
Chen G, Liu Z-P (2022) Graph attention network for link prediction of gene regulations from single-cell RNA-sequencing data. Bioinformatics 38(19):4522–4529. https://doi.org/10.1093/bioinformatics/btac559
Hyvärinen A, Oja E (2000) Independent component analysis: algorithms and applications. Neural Netw 13(4):411–430. https://doi.org/10.1016/s0893-6080(00)00026-5
Kingma DP, Welling M (2013) Auto-encoding variational bayes. arXiv. http://arxiv.org/abs/1312.6114
Zhang Z, Xu J, Wu Y et al (2022) CapsNet-LDA: predicting lncRNA-disease associations using attention mechanism and capsule network based on multi-view data. Brief Bioinform 24(1):bbac531. https://doi.org/10.1093/bib/bbac531
Sun F, Sun J, Zhao Q (2022) A deep learning method for predicting metabolite-disease associations via graph neural network. Brief Bioinform 23(4):bbac266. https://doi.org/10.1093/bib/bbac266
Liu X, Song C, Huang F et al (2022) GraphCDR: a graph neural network method with contrastive learning for cancer drug response prediction. Brief Bioinform 23(1):bbab457. https://doi.org/10.1093/bib/bbab457
Wang H, Huang F, Xiong Z et al (2022) A heterogeneous network-based method with attentive meta-path extraction for predicting drug-target interactions. Brief Bioinform 23(4):bbac184. https://doi.org/10.1093/bib/bbac184
Fu H, Huang F, Liu X et al (2021) MVGCN: data integration through multi-view graph convolutional network for predicting links in biomedical bipartite networks. Bioinformatics 38(2):426–434. https://doi.org/10.1093/bioinformatics/btab651
Wang W, Zhang L, Sun J et al (2022) Predicting the potential human lncRNA-miRNA interactions based on graph convolution network with conditional random field. Brief Bioinform 23(6):bbac463. https://doi.org/10.1093/bib/bbac463
Yi H-C, You Z-H, Huang D-S et al (2022) Graph representation learning in bioinformatics: trends, methods and applications. Brief Bioinform 23(1):bbab340–bbab340. https://doi.org/10.1093/bib/bbab340
Xu K, Hu W, Leskovec J et al (2018) How Powerful are Graph Neural Networks? arXiv. https://doi.org/10.48550/arXiv.1810.00826
Hamilton W, Ying Z, Leskovec J (2017) Inductive representation learning on large graphs. arXiv. https://doi.org/10.48550/arXiv.1706.02216
Peng L, Tan J, Xiong W et al (2023) Deciphering ligand-receptor-mediated intercellular communication based on ensemble deep learning and the joint scoring strategy from single-cell transcriptomic data. Comput Biol Med 163:107137. https://doi.org/10.1016/j.compbiomed.2023.107137
Peng L, Wang F, Wang Z et al (2022) Cell-cell communication inference and analysis in the tumour microenvironments from single-cell transcriptomics: data resources and computational strategies. Brief Bioinform 23(4):bbac234. https://doi.org/10.1093/bib/bbac234
Peng L, Xiong W, Han C et al (2023) Cell Dialog: a computational framework for ligand-receptor-mediated cell-cell communication analysis III. IEEE J Biomed Health Inform 28(1):580–591. https://doi.org/10.1109/jbhi.2023.3333828
Peng L, Tan J, Tian X et al (2022) EnANNDeep: An ensemble-based lncRNA–protein interaction prediction framework with adaptive k-nearest neighbor classifier and deep models. Interdiscip Sci: Comput Life Sci 14(1):209–232. https://doi.org/10.1007/s12539-021-00483-y
Peng L, Yuan R, Han C et al (2023) CellEnBoost: a boosting-based ligand-receptor interaction identification model for cell-to-cell communication inference. IEEE Trans Nanobiosci 22(4):705–715. https://doi.org/10.1109/TNB.2023.3278685
Shen L, Liu F, Huang L et al (2022) VDA-RWLRLS: An anti-SARS-CoV-2 drug prioritizing framework combining an unbalanced bi-random walk and Laplacian regularized least squares. Comput Biol Med 140:105119. https://doi.org/10.1016/j.compbiomed.2021.105119
Chu L-F, Leng N, Zhang J et al (2016) Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol 17(1):173. https://doi.org/10.1186/s13059-016-1033-x
Camp JG, Sekine K, Gerber T et al (2017) Multilineage communication regulates human liver bud development from pluripotency. Nature 546(7659):533–538. https://doi.org/10.1038/nature22796
Shalek AK, Satija R, Shuga J et al (2014) Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510(7505):363–369. https://doi.org/10.1038/nature13437
Hayashi T, Ozaki H, Sasagawa Y et al (2018) Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat Commun 9(1):619. https://doi.org/10.1038/s41467-018-02866-0
Nestorowa S, Hamey FK, Pijuan Sala B et al (2016) A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128(8):e20–e31. https://doi.org/10.1182/blood-2016-05-716480
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Garcia-Alonso L, Holland CH, Ibrahim MM et al (2019) Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 29(8):1363–1375. https://doi.org/10.1101/gr.240663.118
Liu Z-P, Wu C, Miao H et al (2015) RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database 2015(2015):bav095. https://doi.org/10.1093/database/bav095
Han H, Cho J-W, Lee S et al (2018) TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 46(D1):D380–D386. https://doi.org/10.1093/nar/gkx1013
Oki S, Ohta T, Shioi G et al (2018) Ch IP-Atlas: a data-mining suite powered by full integration of public Ch IP-seq data. EMBO Rep 19(12):e46255. https://doi.org/10.15252/embr.201846255
Xu H, Baroukh C, Dannenfelser R et al (2013) ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database 2013(2013):bat045. https://doi.org/10.1093/database/bat045
Moore JE, Purcaro MJ, Pratt HE et al (2020) Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583(7818):699–710. https://doi.org/10.1038/s41586-020-2493-4
Mora-Bermúdez F, Badsha F, Kanton S et al (2016) Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife 5:e18683. https://doi.org/10.7554/eLife.18683
Peng L, Yang C, Chen Y et al (2023) Predicting CircRNA-Disease associations via feature convolution learning with heterogeneous graph attention network. IEEE J Biomed Health Inform 27(6):3072-3082. https://doi.org/10.1109/jbhi.2023.3260863
Zhou Z, Zhuo L, Fu X et al (2024) Joint deep autoencoder and subgraph augmentation for inferring microbial responses to drugs. Brief Bioinform 25(1):bbad483. https://doi.org/10.1093/bib/bbad483
Zhou Z, Zhuo L, Fu X et al (2023) Joint masking and self-supervised strategies for inferring small molecule-miRNA associations. Mol Ther - Nucleic Acids 35:102103. https://doi.org/10.1016/j.omtn.2023.102103
Liu W, Tang T, Lu X et al (2023) MPCLCDA: predicting circRNA–disease associations by using automatically selected meta-path and contrastive learning. Brief Bioinform 24(4):bbad227. https://doi.org/10.1093/bib/bbad227
Liu W, Lin H, Huang L et al (2022) Identification of miRNA–disease associations via deep forest ensemble learning based on autoencoder. Brief Bioinform 23(3):bbac104. https://doi.org/10.1093/bib/bbac104
Junlin X, Jielin X, Yajie M et al (2023) Graph embedding and Gaussian mixture variational autoencoder network for end-to-end analysis of single-cell RNA sequencing data. Cell Rep Methods 3(1):100382. https://doi.org/10.1016/j.crmeth.2022.100382
Tang J, Qu M, Wang M et al (2015) Line: Large-scale information network embedding. In:Proceedings of the 24th international conference on world wide web. https://doi.org/10.1145/2736277.2741093
Higgins I, Matthey L, Pal A et al. beta-vae: Learning basic visual concepts with a constrained variational framework. In: International conference on learning representations. https://openreview.net/forum?id=Sy2fzU9gl
Funding
The Scientific Research Fund of Hunan Provincial Education Department, 22A0101, Wei Liu
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, W., Teng, Z., Li, Z. et al. CVGAE: A Self-Supervised Generative Method for Gene Regulatory Network Inference Using Single-Cell RNA Sequencing Data. Interdiscip Sci Comput Life Sci (2024). https://doi.org/10.1007/s12539-024-00633-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12539-024-00633-y