Abstract
A complete RNA-Seq analysis involves the use of several different tools, with substantial software and computational requirements. The Galaxy platform simplifies the execution of such bioinformatics analyses by embedding the needed tools in its web interface, while also providing reproducibility. Here, we describe how to perform a reference-based RNA-Seq analysis using Galaxy, from data upload to visualization and functional enrichment analysis of differentially expressed genes.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Galaxy
- Workflow
- Visualizations
- Quality control
- Sequence mapping
- Differential gene expression
- Functional enrichment
1 Introduction
In recent years, RNA sequencing (in short RNA-Seq) has become a very widely used technology to analyze the continuously changing cellular transcriptome, that is, the set of all RNA molecules in one cell or a population of cells. One of the most common aims of RNA-Seq is the profiling of gene expression by identifying genes or molecular pathways that are differentially expressed (DE) between two or more biological conditions.
The computational workflow for the detection of DE genes and pathways from RNA-Seq data requires the use of several command-line tools and substantial computational resources that most users may not have access to.
Galaxy [1] is a powerful and easy to use web-based platform for scientific data analysis. Steps in an analysis are executed by running Galaxy tools, which describe how to translate parameters for command-line software into a user-friendly web interface.
The graphical web interface and a large amount of high-quality, community-developed and maintained tools and training materials enable rapid interactive analyses for novices and expert users alike.
For each step in an analysis, Galaxy captures several metadata (e.g., tool identifier and version, inputs, and parameters) enabling reproducibility. Galaxy also allows users to easily share their workflows and data.
Galaxy’s backend architecture can interface with various cloud and high-performance computing (HPC) environments, thereby providing the necessary computing resources to run computationally demanding analyses, while the end users only need access to a web browser.
Galaxy is free, open source software and can be installed locally or used on more than 120 publicly available servers. Galaxy is supported by a large community of users and developers. An important community-maintained resource is the Galaxy Training Material (available at https://training.galaxyproject.org) [2], which hosts a wide range of step-by-step hands-on tutorials for common bioinformatic analysis tasks. In particular, this chapter is based on the “Reference-based RNA-Seq data analysis” tutorial (https://training.galaxyproject.org/topics/transcriptomics/tutorials/ref-based/tutorial.html), and we defer the reader to additional explanations there.
In this chapter we will use a selection of Galaxy tools to show step-by-step how to find differentially expressed genes, from data upload to functional enrichment analysis, using real experimental data.
2 Materials
2.1 RNA-Seq Dataset
In the study of [3], the authors identified genes and pathways regulated by the pasilla (ps) gene (the Drosophila melanogaster homologue of the mammalian splicing regulators Nova-1 and Nova-2 proteins) using RNA-Seq data. They depleted the ps gene in D. melanogaster by RNA interference (RNAi). Total RNA was then isolated and used to prepare both single-end and paired-end RNA-Seq libraries for treated (ps-depleted) and untreated samples. These libraries were sequenced to obtain RNA-Seq reads for each sample. The RNA-Seq data for the treated and untreated samples can be compared to identify the effects of the ps gene depletion on gene expression.
In this chapter, we illustrate the analysis of the gene expression data step by step using seven of the original datasets:
-
Four untreated samples: GSM461176, GSM461177, GSM461178, GSM461182.
-
Three treated samples (ps gene depleted by RNAi): GSM461179, GSM461180, GSM461181.
In the first part of this chapter, we will use the files for two out of the seven samples to demonstrate how to calculate read counts (a measure of the gene expression) from FASTQ (https://en.wikipedia.org/wiki/FASTQ_format) files.
2.2 Computational Resources
The entire analysis described in this article can be conducted efficiently on any Galaxy server which has the required tools and reference genome; a list can be found in the “Available on these Galaxies” menu on the “Reference-based RNA-Seq data analysis” tutorial webpage mentioned above. However, to be sure, the authors recommend using the Galaxy Europe server (https://usegalaxy.eu).
3 Methods
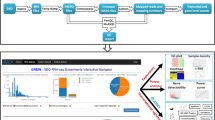
This chapter provides a detailed workflow for the detection of DE genes and gene ontologies from raw RNA-Seq data using Galaxy (Fig. 1). The tutorial starts from quality control of the reads using FastQC and Cutadapt [4]. The reads are then mapped to a reference genome using STAR [5] and checked using the Integrative Genomics Viewer (IGV) [6] and other tools. From the mapped sequences, the number of reads per annotated genes are counted using featureCounts [7]. For each step, quality reports are aggregated using MultiQC [8]. DESeq2 [9] is then used on the read counts to normalize them and extract the differentially expressed genes. Heatmap2 and Volcano Plot are used to visualize DE genes and finally, functional enrichment analysis of the DE genes is performed using goseq [10] to extract interesting Gene Ontologies.
3.1 Upload FASTQ to Galaxy
RNA-Seq analysis usually starts with raw data from the sequencing machine in FASTQ format. Therefore, we first need to upload the FASTQ files for two out of the seven samples into Galaxy.
The Galaxy user interface is split up into four main areas:
-
The top panel for navigating different modes (Analysis, Workflows, Library, Shared Data, User Preferences).
-
The left hand side contains a searchable menu, called the Toolbox, which is used to find and select Tools in the Analysis and Workflow mode.
-
The center panel, whose content changes during the different parts of an analysis. When preparing to run a Galaxy tool or workflow, the user can see and change tool parameters, while it may also be used to show information and metadata for a dataset or its content.
-
The right hand side, called the History, which in the analysis mode shows the list of datasets uploaded or created by previously executed and currently executing tools.
Please login or register for a free account at the Galaxy server you are using to run the tutorial (e.g., https://usegalaxy.eu).
Hands-on: Data Upload
-
1.
Create a new history for this RNA-Seq exercise:
-
(a)
Click the + icon at the top of the history panel.
-
(b)
Click on Unnamed history.
-
(c)
Write a proper name, for example, Reference-based RNA-seq data analysis.
-
(a)
-
2.
Import the FASTQ file pairs from the Shared data library:
-
(a)
Go into Shared Data (top panel) then Data Libraries.
-
(b)
Click on GTN—Material then Transcriptomics, Reference-based RNA-seq data analysis, and https://doi.org/10.5281/zenodo.1185122.
-
(c)
In the search box type fastq to see just the FASTQ files.
-
(d)
Select the following files:
-
https://zenodo.org/api/files/1d804082-4153-47f1-a320-4ac261ce091d/GSM461177_1.fastqsanger
-
https://zenodo.org/api/files/1d804082-4153-47f1-a320-4ac261ce091d/GSM461177_2.fastqsanger
-
https://zenodo.org/api/files/1d804082-4153-47f1-a320-4ac261ce091d/GSM461180_1.fastqsanger
-
https://zenodo.org/api/files/1d804082-4153-47f1-a320-4ac261ce091d/GSM461180_2.fastqsanger
-
-
(e)
Click on the Export to History button near the top and select as Datasets from the drop-down menu.
-
(f)
In the pop-up window, select the history you want to import the files to (or create a new one).
-
(g)
Click on Import.
-
(h)
Click the green pop-up box or Analyze Data in the top panel to move to the analysis page.
-
(a)
-
3.
Rename each dataset according to the sample id (e.g., GSM461177_1):
-
(a)
Click on the pencil icon for the dataset to edit its attributes.
-
(b)
In the central panel, change the Name field.
-
(c)
Click the Save button.
-
(a)
-
4.
Check that the datatype (i.e., format) of each dataset is fastqsanger, not fastq (if needed, click on the dataset name to expand the box to see). If it is not, please change the datatype to fastqsanger.
-
(a)
Click on the pencil icon for the dataset to edit its attributes.
-
(b)
In the central panel, click on the Datatypes tab on the top.
-
(c)
Select fastqsanger.
-
(d)
Click the Change datatype button.
-
(a)
-
5.
Add to each dataset a tag corresponding to the name of the sample (#GSM461177 or #GSM461180):
-
(a)
Click on the dataset.
-
(b)
Click on the Edit dataset tags icon.
-
(c)
Add a tag starting with #. Tags starting with # will be automatically propagated to the outputs of tools using this dataset.
-
(d)
Check that the tag is appearing below the dataset name.
-
(a)
The reads are raw data from the sequencing machine without any preprocessing. They first need to be assessed for their quality.
3.2 Quality Control and Trimming
During sequencing, errors are introduced, such as incorrect nucleotides being called. These are due to the technical limitations of each sequencing platform. Sequencing errors might bias the analysis and can lead to a misinterpretation of the data. Adapters may also be present if the reads are longer than the fragments sequenced and trimming these may improve the number of reads mapped.
Sequence quality control is therefore an essential first step in every analysis. We recommend to use tools such as FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to create a report of sequence quality, MultiQC [8] to aggregate generated reports, and Cutadapt [4] to improve the quality of sequences via trimming and filtering (see Note 1 for alternative tools). Note that to find a tool in Galaxy, you can search for it in the search box at the top of the tool panel on the left. To run a tool after selecting the parameters, just click the Execute button on the tool form.
Hands-on: Quality Control
-
1.
FastQC:
-
(a)
For the “Short read data from your current history” input:
-
Click on the Multiple datasets button.
-
Select all the input datasets you have uploaded by keeping the Ctrl (or COMMAND) key pressed and clicking on the various datasets.
-
-
(a)
-
2.
MultiQC with the following parameters to aggregate the FastQC reports:
-
(a)
In “Results”.
-
“Which tool was used generate logs?”: FastQC.
-
In “FastQC output”.
-
“Type of FastQC output?”: Raw data.
-
“FastQC output”: the 4 RawData files (output of FastQC).
-
-
-
(a)
-
3.
Inspect the web page output from MultiQC for each FASTQ dataset.
The aggregate report shows that everything seems good for three of the files, but in one file (reverse reads of GSM461180) the quality decreases quite a lot at the end of the sequences (Fig. 2).
The “Per base sequence quality” (a) is globally good with a slight decrease at the end of the sequences. For reverse reads of GSM461180, the decrease is quite large. The mean quality score over the reads (b) is quite high, but the distribution is slightly different for reverse reads of GSM461180. All except reverse reads of GSM461180 have a high proportion of duplicated reads (c, expected in RNA-Seq data)
We should trim the reads to get rid of bases that were sequenced with high uncertainty (i.e. low quality bases) at the read ends, and also remove reads of overall bad quality.
Hands-on: Read Trimming and Filtering
-
1.
Cutadapt with the following parameters to trim low quality sequences:
-
(a)
“Single-end or Paired-end reads?”: Paired-end.
-
“FASTQ/A file #1”: both fastqsanger datasets ending with “_1”, selected using the multiple datasets option.
-
“FASTQ/A file #2”: both fastqsanger datasets ending with “_2”, selected using the multiple datasets option.
-
-
(b)
In the “Filter Options” section:
-
“Minimum length”: 20
-
-
(c)
In the “Read Modification Options” section:
-
“Quality cutoff”: 20
-
-
(d)
In the “Output Options” section:
-
“Report”: Yes.
-
-
(a)
-
2.
Inspect the generated Report datasets in your history.
For GSM461177, 5,072,810 bp has been trimmed for the forward reads (read 1) and 8,648,619 bp for the reverse (read 2) because of quality. For GSM461180, 10,224,537 bp for forward and 51,746,850 bp for the reverse. It is not a surprise: we saw that at the end of the reads the quality was dropping more for the reverse reads than for the forward reads, especially for GSM461180.
3.3 Mapping
To make sense of the reads, we need to first figure out where the sequences originated from in the genome, so we can then determine to which genes they belong. When a reference genome for the organism is available, this process is known as aligning or “mapping” reads to the reference.
In this study, the authors used D. melanogaster cells. We should map the quality-controlled sequences to the reference genome of D. melanogaster [11], i.e. the set of nucleic acid sequences assembled as a representative example of the species’ genetic material.
With eukaryotic transcriptomes most reads originate from processed mRNAs lacking introns, therefore they cannot be simply mapped back to the genome as we normally do for DNA data (Fig. 3). Instead, several splice-aware mappers (e.g., TopHat [12], HISAT2 [13, 14], STAR [5]) have been developed to efficiently map transcript-derived reads against a reference genome. Here we will map our reads to the D. melanogaster genome using STAR.
Hands-on: Spliced Mapping
-
1.
Import the Ensembl gene annotation for D. melanogaster (Drosophila_melanogaster.BDGP6.87.gtf) from the Shared Data library into your current Galaxy history.
-
(a)
Rename the dataset if necessary.
-
(b)
Verify that the datatype is gtf and not gff, and that the database is dm6. If not, click on the pencil icon and edit its attributes.
-
(a)
-
2.
RNA STAR to map the reads from both samples on the reference genome:
-
(a)
“Single-end or paired-end reads”: Paired-end (as individual datasets)
-
“RNA-Seq FASTQ/FASTA file, forward reads”: “Read 1 Output” for both samples (outputs of Cutadapt), selected using the multiple datasets option.
-
“RNA-Seq FASTQ/FASTA file, reverse reads”: “Read 2 Output” for both samples (outputs of Cutadapt), selected using the multiple datasets option.
-
-
(b)
“Custom or built-in reference genome”: Use a built-in index.
-
“Reference genome with or without an annotation”: “use genome reference without builtin gene-model”.
-
“Select reference genome”: Fly (Drosophila Melanogaster): “dm6 Full”.
-
“Gene model (gff3,gtf) file for splice junctions”: the imported Drosophila_melanogaster.BDGP6.87.gtf.
-
“Length of the genomic sequence around annotated junctions”: 36 (This parameter should be length of reads—1).
-
-
-
(a)
-
3.
MultiQC to aggregate the STAR logs:
-
(a)
In “Results”.
-
“Which tool was used generate logs?”: STAR.
-
In “STAR output”.
-
“Type of STAR output?”: Log.
-
“STAR output”: log files (outputs of RNA STAR).
-
-
-
(a)
The types of RNA-Seq reads (adapted from Fig. 1a from [13]), reads that mapped entirely within an exon (in red), reads spanning over two exons (in blue), read spanning over more than two exons (in purple)
The MultiQC report reveals that around 80% of reads for both samples are mapped exactly once to the reference genome. Percentages below 70% should be investigated for potential contamination, so here we can safely proceed with the analysis. Both samples have a low (less than 10%) percentage of reads that mapped to multiple locations on the reference genome. This is in the normal range for Illumina short-read sequencing, but the range expected may be lower for long-read sequencing datasets that can span larger repeated regions in the reference sequence.
The main output of STAR is a BAM (https://en.wikipedia.org/wiki/Binary_Alignment_Map) file. The BAM files contain mapping information for all our reads, making it difficult to inspect and explore in text format. A powerful tool to visualize the content of BAM files is the Integrative Genomics Viewer (IGV).
Hands-on: Inspection of Mapping Results
-
1.
Install IGV from https://software.broadinstitute.org/software/igv/download (if not already installed).
-
2.
Start IGV locally.
-
3.
Expand the mapped.bam file (output of RNA STAR) for GSM461177.
-
4.
Click on the local in display with IGV local D. melanogaster (dm6) to load the reads into the IGV browser.
-
5.
IGV: Zoom to chr4:540,000–560,000 (Chromosome 4 between 540 kb to 560 kb) (Fig. 4a).
-
6.
IGV: Inspect the splice junctions using a Sashimi plot (Fig. 4b).
-
(a)
Right click on the BAM file (in IGV).
-
(b)
Select Sashimi Plot from the menu.
-
(a)
Inspection of BAM file with IGV on chromosome 4. (a) On the top, the coverage plot shows the sum of mapped reads at each position as grey peaks. In the middle, each read is displayed where it maps. The blue lines indicate the junction events (or splice sites), that is, reads that are mapped across an intron. On the bottom, the reference genome with its genes is represented. (b) Sashimi plot showing the coverage in red with the arcs representing the splice junctions. The numbers refer to the number of reads spanning the junctions. On the bottom, the different groups of linked boxes represent the different transcripts from the genes at this location that are present in the GTF file
After the mapping, we have now the information on where the reads are located on the reference genome and how well they were mapped (see Note 2 for more quality checks). The next step in RNA-Seq data analysis is quantification of the number of reads mapped to genomic features (genes, transcripts, exons, …). Here we will focus on the genes as we would like to identify the ones that are differentially expressed because of the pasilla gene knockdown.
3.4 Count the Number of Reads per Annotated Genes
To compare the expression of single genes between different conditions (e.g., with or without ps depletion), an essential first step is to quantify the number of reads per gene, or more specifically the number of reads mapping to the exons of each gene. A fast and efficient tool for this task is featureCounts [7] (see Note 3 for alternative tools).
Hands-on: Counting the Number of Reads per Annotated Gene
-
1.
featureCounts to count the number of reads per gene:
-
(a)
“Alignment file”: mapped.bam files (outputs of RNA STAR).
-
(b)
“Specify strand information”: Unstranded (see Note 4).
-
(c)
“Gene annotation file”: in your history.
-
“Gene annotation file”: Drosophila_melanogaster.BDGP6.87.gtf.
-
-
(d)
“Output format”: Gene-ID “\t” read-count (MultiQC/DESeq2/edgeR/limma-voom compatible).
-
(e)
“Create gene-length file”: Yes.
-
(f)
In “Options for paired-end reads”:
-
“Count fragments instead of reads”: Enabled; fragments (or templates) will be counted instead of reads.
-
-
(g)
In “Advanced options”:
-
“GFF feature type filter”: exon.
-
“GFF gene identifier”: gene_id.
-
“Allow reads to map to multiple features”.
-
“Minimum mapping quality per read”: 10
-
-
(a)
-
2.
MultiQC to aggregate the report:
-
(a)
In “Results”:
-
“Which tool was used generate logs?”: featureCounts.
-
“Output of FeatureCounts”: Summary files (outputs of featureCounts).
-
-
(a)
The main output of featureCounts is a table with the number of reads (or fragments in the case of paired-end reads) mapped to each gene (in rows, with their ID in the first column) in the provided annotation. FeatureCounts can also generate a file with the length of each gene, a file we will need later for the functional enrichment analysis.
3.5 Identification of Differentially Expressed Genes
To be able to identify differential gene expression induced by ps depletion, all datasets (three treated and four untreated) must be analyzed following the same procedure. To save time, we have run the previous steps for you and generated seven files with the counts for each gene of D. melanogaster for each sample.
Hands-on: Import all Count Files
-
1.
Create a new empty history.
-
2.
Import the seven count files from the same Shared Data library: GSM461176_untreat_single.counts, GSM461177_untreat_paired.counts, GSM461178_untreat_paired.counts, GSM461179_treat_single.counts, GSM461180_treat_paired.counts, GSM461181_treat_paired.counts, GSM461182_untreat_single.counts.
-
3.
Rename the datasets to the names above (i.e., remove the path prefix).
We would like now to calculate the extent of differential gene expression. DESeq2 [9] is a tool for differential analysis of count data which uses negative binomial generalized linear models (see Note 5 for alternative tools). DESeq2 takes read count files from different samples, combines them into a big table (with genes in the rows and samples in the columns) and applies normalization for sequencing depth and library composition. Gene length normalization does not need to be accounted for because we are comparing the counts between sample groups for the same gene.
DESeq2 also runs the differential gene expression analysis, whose two basic tasks are as follows:
-
Estimate the biological variance using the replicates for each condition (see Note 6 about replicates).
-
Estimate the significance of expression differences between any two conditions.
Multiple factors can be incorporated in the analysis describing known sources of variation (e.g., treatment, tissue type, gender, batches), with two or more levels representing the conditions for each factor. After normalization we can compare the response of the expression of any gene to the presence of different levels of a factor in a statistically reliable way.
In our example, we have samples with two varying factors that can contribute to differences in gene expression: Treatment (either treated or untreated) and Sequencing type (paired-end or single-end). Here, treatment is the primary factor that we are interested in. The sequencing type is further information we know about the data that might affect the analysis. Multifactor analysis allows us to assess the effect of the treatment, while taking the sequencing type into account too.
Hands-on: Determine Differentially Expressed Features
-
1.
DESeq2 with the following parameters:
-
(a)
“how”: Select datasets per level.
-
In “1: Factor”.
-
“Specify a factor name”: Treatment
-
In “Factor level”:
-
1.
In “1: Factor level”:
(a) “Specify a factor level”: treated.
(b) “Counts file(s)”: the three gene count files with
“treat” in their name.
-
2.
In “2: Factor level”:
(a) “Specify a factor level”: untreated
(b) “Counts file(s)”: the four gene count files with
“untreat” in their name.
-
1.
-
-
Click on “Insert Factor” (not on “Insert Factor level”).
-
In “2: Factor”.
-
“Specify a factor name”: Sequencing
-
In “Factor level”:
-
1.
In “1: Factor level”:
(a) “Specify a factor level”: PE
(b) “Counts file(s)”: the four gene count files with
“paired” in their name.
-
2.
In “2: Factor level”:
(a) “Specify a factor level”: SE
(b) “Counts file(s)”: the three gene count files with
“single” in their name.
-
1.
-
-
-
(b)
“Files have header?”: No.
-
(c)
“Visualising the analysis results”: Yes.
-
(d)
“Output normalized counts table”: Yes.
-
(a)
DESeq2 generated three outputs. The first output is the table with the normalized counts for each gene (rows) in the samples (columns). The second output is a graphical summary of the results, useful to evaluate the quality of the experiment:
-
Plot with the first two dimensions from a principal component analysis (PCA) run on the normalized counts of the samples (Fig. 5a). It shows the samples in the 2D plane spanned by their first two principal components. Each replicate is plotted as an individual data point. This type of plot is useful for visualizing the overall effect of experimental covariates and batch effects.
-
Heatmap of the sample-to-sample distance matrix (with clustering) based on the normalized counts (Fig. 5b). The heatmap gives an overview of similarities and dissimilarities between samples: the color represents the distance between the samples. Dark blue means shorter distance, that is, closer samples given the normalized counts.
-
Dispersion estimates: gene-wise estimates (black), the fitted values (red), and the final maximum a posteriori estimates used in testing (blue). This dispersion plot is typical, with the final estimates shrunk from the gene-wise estimates toward the fitted estimates. Some gene-wise estimates are flagged as outliers and not shrunk toward the fitted value. The amount of shrinkage can be more or less than seen here, depending on the sample size, the number of coefficients, the row mean and the variability of the gene-wise estimates.
-
Histogram of p-values for the genes in the comparison between the two levels of the first factor.
-
MA plot. It displays the global view of the relationship between the expression change of conditions (log ratios, M), the average expression strength of the genes (average mean, A), and the ability of the algorithm to detect differential gene expression. The genes that passed the significance threshold (adjusted p-value <0.1) are colored in red.
Graphical summary of DESeq2 results. (a) Plot with the first 2 dimensions from a principal component analysis (PCA) run on the normalized counts of the samples. The first dimension is separating the treated samples from the untreated samples and the second dimension the single-end datasets from the paired-end datasets. The datasets are grouped following the levels of the two factors. No hidden effect seems to be present on the data. If there is unwanted variation present in the data (e.g., batch effects) it is always recommended to correct for this, which can be accommodated in DESeq2 by including in the design any known batch variables. (b) Heatmap of the sample-to-sample distance matrix (with clustering) based on the normalized counts. The samples are first grouped by the treatment (the first factor) and secondly by the sequencing type (the second factor), as in the PCA plot
The main output of DESeq2 is a summary file with the following values for each gene:
-
Gene identifier.
-
Mean normalized counts, averaged overall samples from both conditions.
-
Fold change in log2 (logarithm base 2). The log2 fold changes are based on the primary factor level 1 vs factor level 2, hence the input order of factor levels is important. Here, DESeq2 computes fold changes of ‘treated’ samples against ‘untreated’ from the first factor ‘Treatment’, i.e. the values correspond to up- or downregulation of genes in treated samples (see Note 7 for details about other factors and levels comparisons).
-
Standard error estimate for the log2 fold change estimate.
-
Wald statistic.
-
p-Value for the statistical significance of this change,
-
p-Value adjusted for multiple testing with the Benjamini–Hochberg procedure, which controls the false discovery rate (FDR).
For example, the gene FBgn0003360 is differentially expressed because of the treatment: it has a significant adjusted p-value (4.0 × 10−178, much less than 0.05) and it is less expressed (− in the log2FC column) in treated samples compared to untreated samples, by a factor ~8 (2|log2FC|).
Some of the tools we will use in the rest of the chapter require a header row in the DESeq2 result file so we will add column names before going further.
Hands-on: Add Column Names
-
1.
Create a new file from the following (header line of the DESeq2 output) by pasting the line below into the Galaxy upload file Paste/Fetch data box:
GeneID Base-mean log2(FC) StdErr Wald-Stats P-value P-adj
-
(a)
In Type, select “Tabular”.
-
(b)
In Settings, click on “Convert spaces to tabs”.
-
(a)
-
2.
Concatenate datasets to add the header line to the annotated genes.
-
(a)
“Concatenate”: the Pasted entry dataset.
-
(b)
“Dataset”: the DESeq2 result file.
-
(a)
We would like to extract the most differentially expressed genes due to the treatment and with an absolute fold change >2 (equivalent to an absolute log2FC > 1).
Hands-on: Extract the most Differentially Expressed Genes
-
1.
Filter data on any column using simple expressions to extract genes with a significant change in gene expression (adjusted p-value below 0.05) between treated and untreated samples:
-
(a)
“Filter”: output of Concatenate.
-
(b)
“With following condition”: c7 < 0.05
-
(c)
“Number of header lines to skip”: 1
-
(a)
-
2.
Rename the output “Genes with significant adj p-value”.
-
3.
Filter data on any column using simple expressions to extract genes with an absolute fold change (FC) > 2.
-
(a)
“Filter”: Genes with significant adj p-value.
-
(b)
“With following condition”: abs(c3) > 1
-
(c)
“Number of header lines to skip”: 1
-
(a)
We now have a table with 131 lines corresponding to the most differentially expressed genes. For each gene, we have its ID, its mean normalized counts (averaged overall samples from both conditions), its log2FC and other information.
The ID for each gene is something like FBgn0003360, which is an ID from the corresponding database, here Flybase [15]. These IDs are unique but sometimes we prefer to have the gene symbols, even if they may not reference a unique gene (e.g., duplicated after reannotation), as they may hint already to a function or they help you to search for desired candidates. We would also like to display the location of these genes on the genome. We can extract such information from the annotation file which we used for mapping and counting.
Hands-on: Annotate the Differentially Expressed Genes
-
1.
Using View all histories, drag and drop the Ensembl gene annotation for D. melanogaster (Drosophila_melanogaster.BDGP6.87.gtf) from the previous history into this history.
-
2.
Annotate DESeq2/DEXSeq output tables with:
-
(a)
“Tabular output of DESeq2/edgeR/limma/DEXSeq”: output of the last Filter.
-
(b)
“Input file type”: DESeq2/edgeR/limma.
-
(c)
“Reference annotation in GFF/GTF format”: Drosophila_melanogaster.BDGP6.87.gtf.
-
(a)
The generated output is an extension of the previous file: (1) Gene identifiers, (2) Mean normalized counts overall samples, (3) Log2 fold change, (4) Standard error estimate for the log2 fold change estimate, (5) Wald statistic, (6) p-value for the Wald statistic, (7) p-value adjusted for multiple testing with the Benjamini–Hochberg procedure for the Wald statistic, (8) Chromosome, (9) Start, (10) End, (11) Strand, (12) Feature, (13) Gene name.
With this extra information, we can see that FBgn0025111, one of the most significantly overexpressed genes, is located on the reverse strand of chromosome X, between 10,778,953 bp and 10,786,907 bp, and is also named Ant2, that is, that it corresponds to adenine nucleotide translocase 2.
3.6 Visualization
We can visualize the differentially expressed results with volcano plots and heatmaps.
We can generate a heatmap of expression for the top differentially expressed genes in the different samples. To do this we need the normalized counts for these genes. To extract the normalized counts for the interesting genes, we join the normalized count table generated by DESeq2 with the table of the top differentially expressed genes that we just generated. We can then use heatmap2 to create the heatmap. In heatmap2 we will select to scale the data by row (genes), which converts the expression values to z-scores and prevents highly expressed genes from dominating the plot. However, note that heatmap2 performs clustering before scaling, so if you want to view the clustering after scaling, use the Table Compute tool to compute Z-scores before creating the heatmap.
Hands-on: Create an Expression Heatmap for the Top Differentially Expressed Genes
-
1.
Join two Datasets side by side on a specified field to keep only the most differentially expressed genes in the DESeq2 normalized counts file:
-
(a)
“Join”: the DESeq2 normalized counts file.
-
(b)
“using column”: Column: 1.
-
(c)
“with”: output from Annotate DESeq2/DEXSeq.
-
(d)
“and column”: Column: 1.
-
(e)
“Keep lines of first input that do not join with second input”: No.
-
(f)
“Keep the header lines”: Yes.
-
(a)
-
2.
Cut columns from a table to extract the columns with the gene IDs and normalized counts:
-
(a)
“Cut columns”: c1-c8
-
(b)
“Delimited by”: Tab.
-
(c)
“From”: the output of the previous Join.
-
(a)
-
3.
heatmap2 to create a heatmap:
-
(a)
“Input should have column headers - these will be the columns that are plotted”: the file from the previous Cut.
-
(b)
“Plot title”: Top differentially expressed genes
-
(c)
“Data transformation”: Log2(value) transform my data.
-
(d)
“Enable data clustering”: Yes.
-
(e)
“Clustering columns and rows”: Cluster rows and columns.
-
(f)
“Labeling columns and rows”: Label columns and not rows.
-
(g)
“Coloring groups”: Blue to white to red.
-
(h)
“Data scaling”: Scale my data by row.
-
(a)
Based on the normalized counts for the 130 top differentially expressed genes, the samples (in columns) are clustered primarily by treatment (Fig. 6a). We can see that the samples within each treatment type (treated and untreated) tend to have similar expression patterns for these genes (low expression is blue and high expression is red), which is good. We can see also clusters of genes based on their expression.
Visualization of the expression results. (a) Heatmap of normalized expression (z-scores) for the top 130 differentially expressed genes in the 7 samples. Blue indicates relatively low expression in a sample, red indicates high. (b) Volcano plot for the comparison between treated and untreated samples, showing all genes, with log2FC on the X-axis and −log10 of the P-value on the Y-axis. The points in grey correspond to genes that are not significantly differentially expressed (using a threshold of 0.05 on the adjusted p-value and absolute log2FC of 1), in red the significantly overexpressed genes (log2FC > 1) and in blue the significantly underexpressed genes (log2FC < −1)
Volcano plots are commonly used to display the results of RNA-Seq or other omics experiments. A volcano plot is a type of scatterplot that shows statistical significance (P value) versus magnitude of change (fold change). It enables quick visual identification of genes with large fold changes that are also statistically significant. These may be the most biologically significant genes. In a volcano plot, the most upregulated genes are toward the right, the most downregulated genes are toward the left, and the most statistically significant genes are toward the top. We will make a volcano plot showing the names of the top ten most differentially expressed genes.
Hands-on: Creating a Volcano Plot
-
1.
Filter data on any column using simple expressions to remove genes with NAs from the DESeq2 result:
-
(a)
“Filter”: output from Concatenate.
-
(b)
“With following condition”: c7!='NA'
-
(c)
“Number of header lines to skip”: 1
-
(a)
-
2.
Join two Datasets side by side on a specified field to add the gene names for the most differentially expressed genes to the DESeq2 results file:
-
(a)
“Join”: output from the previous Filter.
-
(b)
“using column”: Column: 1.
-
(c)
“with”: output from Annotate DESeq2/DEXSeq.
-
(d)
“and column”: Column: 1.
-
(e)
“Keep lines of first input that do not join with second input”: Yes.
-
(f)
“Keep the header lines”: Yes.
-
(a)
-
3.
Volcano Plot to create a volcano plot:
-
(a)
“Specify an input file”: output of the previous Join.
-
(b)
“FDR (adjusted P value)”: Column: 7.
-
(c)
“P value (raw)”: Column: 6.
-
(d)
“Log Fold Change”: Column: 3.
-
(e)
“Labels”: Column: 20.
-
(f)
“Significance threshold”: 0.05
-
(g)
“LogFC threshold to colour”: 1.0
-
(h)
“Points to label”: Significant.
-
“Only label top most significant”: 10
-
-
(i)
In “Plot options”:
-
“Label boxes”: No.
-
-
(a)
Figure. 6b shows a volcano plot for this dataset. The significantly differentially expressed genes (using thresholds of adjusted p-value 0.05 and absolute log2FC of 1) are colored red if they are upregulated and blue if they are downregulated. The top ten most significantly differentially expressed genes by P value are labeled. In this plot we can see that the most significantly upregulated gene is Ant2, the most downregulated is Kal1, and that the top ten genes are mostly downregulated (8/10 genes).
3.7 Functional Enrichment Analysis
We have extracted genes that are differentially expressed in treated (ps gene-depleted) samples compared to untreated samples. Now, we would like to know if the differentially expressed genes are enriched transcripts of genes which belong to more common or specific categories in order to identify biological functions that might be impacted. Gene Ontology (GO) analysis is widely used to reduce complexity and highlight biological processes in genome-wide expression studies. To perform the GO analysis of RNA-Seq data, we will use the goseq tool [10]. goseq provides methods for performing GO analysis of RNA-Seq data while taking gene length bias into account. Goseq could also be applied to other category-based tests of RNA-Seq data, such as KEGG pathway analysis.
goseq needs two files as inputs:
-
A tabular file with differentially expressed genes from all genes assayed in the RNA-Seq experiment with two columns: the Gene IDs (unique within the file), in uppercase letters; a Boolean indicating whether the gene is differentially expressed or not (“True” if differentially expressed and “False” if not).
-
A file with information about the length of a gene to correct for potential length bias in differentially expressed genes.
Hands-on: Prepare the Datasets for Goseq
-
1.
Compute an expression on every row with.
-
(a)
“Add expression”: bool(c7 < 0.05)
-
(b)
“as a new column to”: the DESeq2 result file.
-
(a)
-
2.
Cut with.
-
(a)
“Cut columns”: c1,c8
-
(b)
“Delimited by”: Tab.
-
(c)
“From”: the output of the Compute.
-
(a)
-
3.
Change Case with.
-
(a)
“From”: the output of the previous Cut.
-
(b)
“Change case of columns”: c1
-
(c)
“Delimited by”: Tab.
-
(d)
“To”: Upper case.
-
(a)
-
4.
Rename the output to “Gene IDs and differentially expression”.
-
5.
Drag and drop one of the feature length datasets generated by featureCounts into this history using the View all histories.
-
6.
Change Case with.
-
(a)
“From”: the feature lengths (output of featureCounts).
-
(b)
“Change case of columns”: c1
-
(c)
“Delimited by”: Tab.
-
(d)
“To”: Upper case.
-
(a)
-
7.
Rename the output to “Gene IDs and length”.
We now have the two required input files for goseq.
Hands-on: Perform GO Analysis
-
1.
goseq with.
-
(a)
“Differentially expressed genes file”: Gene IDs and differentially expression.
-
(b)
“Gene lengths file”: Gene IDs and length.
-
(c)
“Gene categories”: Get categories.
-
“Select a genome to use”: Fruit fly (dm6).
-
“Select Gene ID format”: Ensembl Gene ID.
-
“Select one or more categories”: GO: Cellular Component, GO: Biological Process, GO: Molecular Function.
-
-
(d)
In “Output Options”.
-
“Output Top GO terms plot?”: Yes.
-
“Extract the DE genes for the categories (GO/KEGG terms)?”: Yes.
-
-
(a)
The main output of goseq is a table (“Ranked category list - Wallenius method”) with the following columns for each GO term:
-
1.
GO category (“category”).
-
2.
p-Value for overrepresentation of the term in the differentially expressed genes (“over_rep_pval”).
-
3.
p-Value for underrepresentation of the term in the differentially expressed genes (under_rep_pval).
-
4.
Number of differentially expressed genes in this category (“numDEInCat”).
-
5.
Number of genes in this category (“numInCat”).
-
6.
Details about the term.
-
7.
Ontology with MF for “Molecular Function” (molecular activities of gene products), CC for “Cellular Component” (where gene products are active), BP for“Biological Process” (pathways and larger processes made up of the activities of multiple gene products).
-
8.
p-Value for overrepresentation of the term in the differentially expressed genes, adjusted for multiple testing with the Benjamini–Hochberg procedure (“p.adjust.over_represented”).
-
9.
p-Value for underrepresentation of the term in the differentially expressed genes, adjusted for multiple testing with the Benjamini–Hochberg procedure (“p.adjust.under_represented”).
From this table we can extract the 31 overrepresented GO terms (using the Filter tool on column 8) and the 83 underrepresented terms (using the Filter tool on column 9), and then group them (using Group data tool on column 7 and count on column 1) to identify that over the 31 overrepresented GO terms, 20 are BP, 3 CC, and 8 MF.
goseq generates also a graph with the top ten overrepresented GO terms and a table with differentially expressed genes (from the list we provided) associated to the GO terms (DE genes for categories (GO/KEGG terms)).
In this chapter, we covered only GO enrichment analysis with goseq, but other gene set enrichment analysis can be done with Galaxy (see Note 8).
3.8 Sharing the Results
Using Galaxy to perform this analysis makes it is both reusable and shareable. In fact, it is possible to simply extract a workflow from a Galaxy history that describes each step of the analysis (tool with parameters used, connections to previous and following steps). This workflow can then be applied on the same or different data, guaranteeing reproducibility.
Moreover, Galaxy histories and workflows can be effortlessly shared with other selected users (via their Galaxy user account or a link), or made publicly available to anyone. See the Galaxy 101 tutorial (https://training.galaxyproject.org/training-material/topics/introduction/tutorials/galaxy-intro-101/tutorial.html) for more details.
4 Conclusion
In this tutorial, we have used the Galaxy platform to perform a complex reference-based RNA-Seq analysis through a web interface in a reproducible and easily shareable way. We extracted meaningful information from the RNA sequencing data, such as which genes are up or downregulated by the depletion of the pasilla gene, and also which GO terms are enriched.
5 Notes
-
1.
As alternative to Cutadapt, the Trim Galore! or Trimmomatic tools can be used.
-
2.
In addition to checking the mapping percentage and quick visual check using IGV, the quality of the data and mapping can be checked further:
-
(a)
Duplicate reads: Duplicate reads can come from highly expressed genes, therefore they are usually kept in RNA-Seq differential expression analysis. But a high percentage of duplicates may indicate an issue, for example, overamplification during PCR of low complexity library. MarkDuplicates from the Picard suite (http://broadinstitute.github.io/picard/) can examine aligned records from a BAM file to locate duplicate reads, that is, reads mapping to the same location (based on the start position of the mapping). In general, up to 50% of duplication can be considered normal to obtain. So both our samples are good.
-
(b)
Number of reads mapped to each chromosome: To assess the sample quality (e.g., excess of mitochondrial contamination), check the sex of samples, or see if any chromosome have highly expressed genes, we can check the numbers of reads mapped to each chromosome using IdxStats from the Samtools suite.
-
(c)
Gene body coverage: The gene body is the different regions of a gene. It is important to check if reads coverage is uniform over gene body or if there is any 5′–3′ bias. For example, a bias toward the 5′ end of genes could indicate degradation of the RNA. Alternatively, a 3′ bias could indicate that the data is from a 3′ assay. To assess this, we can use the Gene Body Coverage tool from the RSeQC tool suite [16]. This tool scales all transcripts to 100 nucleotides (using a provided annotation file) and calculates the number of reads covering each nucleotide position.
-
(d)
Read distribution across features: With RNA-Seq data, we expect most reads to map to exons rather than introns or intergenic regions. Before going further in counting and differential expression analysis, it may be interesting to check the distribution of reads across known gene features (exons, CDS, 5′ UTR, 3′ UTR, introns, intergenic regions). For example, a high number of reads mapping to intergenic regions may indicate the presence of DNA contamination. We can use the Read Distribution tool from the RSeQC tool suite, which uses the annotation file to identify the position of the different gene features.
-
(a)
-
3.
As an alternative to featureCounts, the HTSeq-count [17] tool can be used.
-
4.
RNAs that are typically targeted in RNA-Seq experiments are single stranded (e.g., mRNAs) and thus have polarity (5′ and 3′ ends that are functionally distinct). During a typical RNA-Seq experiment, the information about strandness is lost after both strands of cDNA are synthesized, size selected, and converted into a sequencing library. However, this information can be quite useful for the read counting step, especially for reads located on the overlap of two genes that are on different strands.
Some library preparation protocols create so called stranded RNA-Seq libraries that preserve the strand information. This information can be estimated using a tool called Infer Experiment from the RSeQC [16] tool suite. This tool takes the BAM files from the mapping, selects a subsample of the reads and compares their genome coordinates and strands with those of the reference gene model (from an annotation file). Based on the strand of the genes, it can gauge whether sequencing is strand-specific, and if so, how reads are stranded (forward or reverse).
-
5.
Alternative tools that could be used instead of DESeq2 are edgeR and limma-voom.
-
6.
The expression analysis is estimated from read counts and attempts are made to correct for variability in measurements using replicates, which are absolutely essential for accurate results. For your own analysis, we advise you to use at least 3, but preferably 5, biological replicates per condition. It is possible to have different numbers of replicates per condition.
A technical replicate is an experiment which is performed once but measured several times (e.g., multiple sequencing of the same library). A biological replicate is an experiment performed (and also measured) several times.
In our data, we have four biological replicates (here called samples) without treatment and three biological replicates with treatment (pasilla gene depleted by RNAi).
We recommend to combine the count tables for different technical replicates (but not for biological replicates) before a differential expression analysis.
-
7.
DESeq2 in Galaxy returns the comparison between the different levels for the first factor, after correction for the variability due to the second factor. In our current case, treated against untreated for any sequencing type. To compare sequencing types, we should run DESeq2 again switching factors: factor 1 (treatment) becomes factor 2 and factor 2 (sequencing) becomes factor 1.
To compare the effect of two factors, for example to see if there is a difference in the treatment effect detected with paired vs. single end data, we should run DESeq2 another time but with only one factor with the following four levels: treated-PE, untreated-PE, treated-SE, untreated-SE. By selecting “Output all levels vs all levels of primary factor (use when you have >2 levels for primary factor)” to “Yes,” we can then compare treated-PE vs treated-SE.
-
8.
goseq can also be used to identify interesting pathways by replacing GO terms with KEGG pathways. The KEGG database is a collection of pathway maps representing the current knowledge on the molecular interaction, reaction and relation networks. A map can integrate many entities including genes, proteins, RNAs, chemical compounds, glycans, and chemical reactions, as well as disease genes and drug targets.
From the goseq output, we could investigate which genes are involved in which pathways by looking at the second file generated by goseq. This can be less cumbersome if the pathways are represented as a diagram: Pathview [18] can help to generate automatically pathway representation with information about the genes (e.g., expression).
Other gene set enrichment tools available for Galaxy include fgsea [19] and EGSEA [20]. fgsea (fast gene set enrichment analysis) takes a ranked list of genes and some gene sets to test, such as from the Molecular Signatures Database (MSigDB), and identifies enriched gene sets. It produces a table of enriched gene sets and barcode plots showing the ranking of the gene set. MSigDB only provide gene sets for human, but if you are using another species you could first map the nonhuman gene ids to human. EGSEA (Ensemble of Gene Set Enrichment Analyses) is another gene set enrichment tool that takes a table of counts and built-in gene sets, including MSigDB, and runs a number of enrichment algorithms. It produces a table of enriched gene sets and different types of plots, such as KEGG diagrams. EGSEA provides built-in gene sets for human, mouse and rat, including those from MSigDB.
References
Afgan E, Baker D, Batut B et al (2018) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544
Batut B, Hiltemann S, Bagnacani A et al (2018) Community-driven data analysis training for biology. Cell Syst 6:752–758.e1
Brooks AN, Yang L, Duff MO et al (2011) Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res 21:193–202
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Robinson JT, Thorvaldsdóttir H, Winckler W et al (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930
Ewels P, Magnusson M, Lundin S, Käller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14
dos Santos G, Schroeder AJ, Goodman JL et al (2015) FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res 43:D690–D697
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Kim D, Paggi JM, Park C et al (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915
Thurmond J, Goodman JL, Strelets VB et al (2019) FlyBase 2.0: the next generation. Nucleic Acids Res 47:D759–D765
Wang L, Wang S, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28:2184–2185
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169
Luo W, Brouwer C (2013) Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29:1830–1831
Korotkevich G, Sukhov V, Sergushichev A (2019) Fast gene set enrichment analysis. bioRxiv 060012
Alhamdoosh M, Ng M, Wilson NJ et al (2017) Combining multiple tools outperforms individual methods in gene set enrichment analyses. Bioinformatics 33:414–424
Acknowledgments
This book chapter is based upon the online tutorial “Reference-based RNA-Seq data analysis” (https://training.galaxyproject.org/training-material/topics/transcriptomics/tutorials/ref-based/tutorial.html) © 2019 Bérénice Batut, Mallory Freeberg, Mo Heydarian, Anika Erxleben, Pavankumar Videm, Clemens Blank, Maria Doyle, Nicola Soranzo, Peter van Heusden, licensed under the Creative Commons Attribution 4.0 International Public License (https://creativecommons.org/licenses/by/4.0/).
The authors would like to kindly thank all contributors of the Galaxy Training Material.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this protocol
Cite this protocol
Batut, B., van den Beek, M., Doyle, M.A., Soranzo, N. (2021). RNA-Seq Data Analysis in Galaxy. In: Picardi, E. (eds) RNA Bioinformatics. Methods in Molecular Biology, vol 2284. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1307-8_20
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1307-8_20
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1306-1
Online ISBN: 978-1-0716-1307-8
eBook Packages: Springer Protocols