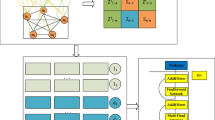
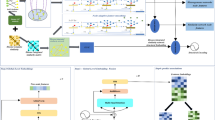
Abstract
Accumulating studies have demonstrated close relationships between long non-coding RNAs (lncRNAs) and diseases. Identification of new lncRNA–disease associations (LDAs) enables us to better understand disease mechanisms and further provides promising insights into cancer targeted therapy and anti-cancer drug design. Here, we present an LDA prediction framework called GEnDDn based on deep learning. GEnDDn mainly comprises two steps: First, features of both lncRNAs and diseases are extracted by combining similarity computation, non-negative matrix factorization, and graph attention auto-encoder, respectively. And each lncRNA–disease pair (LDP) is depicted as a vector based on concatenation operation on the extracted features. Subsequently, unknown LDPs are classified by aggregating dual-net neural architecture and deep neural network. Using six different evaluation metrics, we found that GEnDDn surpassed four competing LDA identification methods (SDLDA, LDNFSGB, IPCARF, LDASR) on the lncRNADisease and MNDR databases under fivefold cross-validation experiments on lncRNAs, diseases, LDPs, and independent lncRNAs and independent diseases, respectively. Ablation experiments further validated the powerful LDA prediction performance of GEnDDn. Furthermore, we utilized GEnDDn to find underlying lncRNAs for lung cancer and breast cancer. The results elucidated that there may be dense linkages between IFNG-AS1 and lung cancer as well as between HIF1A-AS1 and breast cancer. The results require further biomedical experimental verification. GEnDDn is publicly available at https://github.com/plhhnu/GEnDDn.
Graphic Abstract









Similar content being viewed by others

Availability of Data and Materials
Datasets and codes are publicly available at https://github.com/plhhnuGEnDDn.
Code Availability
Datasets and codes are publicly available at https://github.com/plhhnu/GEnDDn.
References
Ma L, Zhang Z (2023) The contribution of databases towards understanding the universe of long non-coding RNAs. Nat Rev Mol Cell Biol 24(9):601–602. https://doi.org/10.1038/s41580-023-00612-z
Sheng N, Huang L, Lu Y et al (2023) Data resources and computational methods for lncRNA–disease association prediction. Comput Biol Med 153:106527. https://doi.org/10.1016/j.compbiomed.2022.106527
Peng L, Tan J, Tian X et al (2022) EnANNDeep: an ensemble-based lncRNA–protein interaction prediction framework with adaptive k-nearest neighbor classifier and deep models. Interdiscip Sci 14(1):209–232. https://doi.org/10.1007/s12539-021-00483-y
Zhang Z, Xu J, Wu Y et al (2023) CapsNet-LDA: predicting lncRNA-disease associations using attention mechanism and capsule network based on multi-view data. Brief Bioinform 24(1):bbac531. https://doi.org/10.1093/bib/bbac531
Guo H, Zhao T (2023) Multiple mutations of IFITM3 are associated with COVID-19 susceptibilityregulation function of IFITM3 in COVID-19. J Infect 86(6):614–616. https://doi.org/10.1016/j.jinf.2023.02.032
Chen X, Huang L (2023) Computational model for disease research. Brief Bioinform 24(1):bbac615. https://doi.org/10.1093/bib/bbac615
Wang W, Zhang L, Sun J et al (2022) Predicting the potential human lncRNA–miRNA interactions based on graph convolution network with conditional random field. Brief Bioinform 23(6):bbac463. https://doi.org/10.1093/bib/bbac463
Chen X, Yan CC, Zhang X et al (2017) Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform 18(4):558–576. https://doi.org/10.1093/bib/bbw060
Chen X, Sun Y, Guan N et al (2019) Computational models for lncRNA function prediction and functional similarity calculation. Brief Funct Genom 18(1):58–82. https://doi.org/10.1093/bfgp/ely031
Xie G-B, Chen R-B, Lin Z et al (2023) Predicting lncRNA–disease associations based on combining selective similarity matrix fusion and bidirectional linear neighborhood label propagation. Brief Bioinform 24(1):bbac595. https://doi.org/10.1093/bib/bbac595
Li X, Jin F, Li Y (2021) A novel autophagy-related lncRNA prognostic risk model for breast cancer. J Cell Mol Med 25(1):4–14. https://doi.org/10.1111/jcmm.15980
Liu S, Sun Y, Hou Y et al (2021) A novel lncRNA ROPM-mediated lipid metabolism governs breast cancer stem cell properties. J Hematol Oncol 14:1–23. https://doi.org/10.1186/s13045-021-01194-z
Wu Z, Lu Z, Li L et al (2022) Identification and validation of ferroptosis-related LncRNA signatures as a novel prognostic model for colon cancer. Front Immunol 12:783362. https://doi.org/10.3389/fimmu.2021.783362
Hu Q, Ma H, Chen H et al (2022) LncRNA in tumorigenesis of non-small-cell lung cancer: from bench to bedside. Cell Death Discov 8(1):359. https://doi.org/10.1038/s41420-022-01157-4
He Y, Jiang X, Duan L et al (2021) LncRNA PKMYT1AR promotes cancer stem cell maintenance in non-small cell lung cancer via activating Wnt signaling pathway. Mol Cancer 20(1):1–21. https://doi.org/10.1186/s12943-021-01469-6
Chen X, Huang L (2022) Computational model for ncRNA research. Brief Bioinform 23(6):bbac472. https://doi.org/10.1093/bib/bbac472
Wang H, Tang J, Ding Y et al (2021) Exploring associations of non-coding RNAs in human diseases via three-matrix factorization with hypergraph-regular terms on center kernel alignment. Brief Bioinform 22(5):bbaa409. https://doi.org/10.1093/bib/bbaa409
Zhao J, Zhao B, Song X et al (2023) Subtype-DCC: decoupled contrastive clustering method for cancer subtype identification based on multi-omics data. Brief Bioinform 24(2):bbad025. https://doi.org/10.1093/bib/bbad025
Chen X, Sun L, Zhao Y (2021) NCMCMDA: miRNA–disease association prediction through neighborhood constraint matrix completion. Brief Bioinform 22(1):485–496. https://doi.org/10.1093/bib/bbz159
Wu J, Zhao X, Zhao X et al (2023) Multi-view contrastive heterogeneous graph attention network for lncRNA–disease association prediction. Brief Bioinform 24(1):bbac548. https://doi.org/10.1093/bib/bbac548
Chen X, Yan G (2013) Novel human lncRNA–disease association inference based on lncRNA expression profiles. Bioinformatics 29(20):2617–2624. https://doi.org/10.1093/bioinformatics/btt426
Lu C, Yang M, Li M et al (2019) Predicting human lncRNA–disease associations based on geometric matrix completion. IEEE J Biomed Health 24(8):2420–2429. https://doi.org/10.1109/JBHI.2019.2958389
Xie G, Huang B, Sun Y et al (2021) RWSF-BLP: a novel lncRNA–disease association prediction model using random walk-based multi-similarity fusion and bidirectional label propagation. Mol Genet Genom 296:473–483. https://doi.org/10.1007/s00438-021-01764-3
Xie G, Zhu Y, Lin Z et al (2022) HBRWRLDA: predicting potential lncRNA–disease associations based on hypergraph bi-random walk with restart. Mol Genet Genom 297(5):1215–1228. https://doi.org/10.1007/s00438-022-01909-y
Zhang G, Gao Y (2023) BRWMC: predicting lncRNA–disease associations based on bi-random walk and matrix completion on disease and lncRNA networks. Comput Biol Chem 103:107833. https://doi.org/10.1016/j.compbiolchem.2023.107833
Xie G-B, Liu S, Gu G-S et al (2023) LUNCRW: prediction of potential lncRNA–disease associations based on unbalanced neighborhood constraint random walk. Anal Biochem 679:115297. https://doi.org/10.1016/j.ab.2023.115297
Huang L, Zhang L, Chen X (2022) Updated review of advances in microRNAs and complex diseases: taxonomy, trends and challenges of computational models. Brief Bioinform 23(5):bbac358. https://doi.org/10.1093/bib/bbac358
Yu G, Wang Y, Wang J et al (2020) Attributed heterogeneous network fusion via collaborative matrix tri-factorization. Inform Fusion 63:153–165. https://doi.org/10.1016/j.inffus.2020.06.012
Qiu S, Wang M, Yang Y et al (2023) Meta multi-instance multi-label learning by heterogeneous network fusion. Inform Fusion 94:272–283. https://doi.org/10.1016/j.inffus.2023.02.010
Jiang Q, Ma R, Wang J et al (2015) LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. In: BMC genomics, vol 16, no 3. BioMed Central, pp 1–11. https://doi.org/10.1186/1471-2164-16-S3-S2
Feng H, Jin D, Li J et al (2023) Matrix reconstruction with reliable neighbors for predicting potential MiRNA–disease associations. Brief Bioinform 24(1):bbac571. https://doi.org/10.1093/bib/bbac571
Chu Y, Wang X, Dai Q et al (2021) MDA-GCNFTG: identifying miRNA–disease associations based on graph convolutional networks via graph sampling through the feature and topology graph. Brief Bioinform 22(6):bbab165. https://doi.org/10.1093/bib/bbab165
Chen X, Xie D, Zhao Q et al (2019) MicroRNAs and complex diseases: from experimental results to computational models. Brief Bioinform 20(2):515–539. https://doi.org/10.1093/bib/bbx130
Fu G, Wang J, Domeniconi C et al (2018) Matrix factorization-based data fusion for the prediction of lncRNA–disease associations. Bioinformatics 34(9):1529–1537. https://doi.org/10.1093/bioinformatics/btx794
Yu J, Xuan Z, Feng X et al (2019) A novel collaborative filtering model for LncRNA–disease association prediction based on the Naïve Bayesian classifier. BMC Bioinform 20:1–13. https://doi.org/10.1186/s12859-019-2985-0
Wang Y, Yu G, Wang J et al (2020) Weighted matrix factorization on multi-relational data for LncRNA–disease association prediction. Methods 173:32–43. https://doi.org/10.1016/j.ymeth.2019.06.015
Wang Y, Yu G, Domeniconi C et al (2019) Selective matrix factorization for multi-relational data fusion. In: International conference on database systems for advanced applications. Springer, Berlin, pp 313–329. https://doi.org/10.1007/978-3-030-18576-3_19
Zhu R, Wang Y, Liu J et al (2021) IPCARF: improving lncRNA–disease association prediction using incremental principal component analysis feature selection and a random forest classifier. BMC Bioinform 22(1):1–17. https://doi.org/10.1186/s12859-021-04104-9
Li J, Li J, Kong M et al (2021) SVDNVLDA: predicting lncRNA–disease associations by singular value decomposition and node2vec. BMC Bioinform 22:1–18. https://doi.org/10.1186/s12859-021-04457-1
Lu C, Xie M (2023) LDAEXC: LncRNA–disease associations prediction with deep autoencoder and XGBoost classifier. Interdiscip Sci 15(3):439–451. https://doi.org/10.1007/s12539-023-00573-z
Wu H, Liang Q, Zhang W et al (2022) iLncDA-LTR: identification of lncRNA–disease associations by learning to rank. Comput Biol Med 146:105605. https://doi.org/10.1016/j.compbiomed.2022.105605
Liang Q, Zhang W, Wu H et al (2023) LncRNA–disease association identification using graph auto-encoder and learning to rank. Brief Bioinform 24(1):bbac539. https://doi.org/10.1093/bib/bbac539
Peng L, Huang L, Su Q et al (2024) LDA-VGHB: identifying potential lncRNA–disease associations with singular value decomposition, variational graph auto-encoder and heterogeneous Newton boosting machine. Brief Bioinform 25(1):bbad466. https://doi.org/10.1093/bib/bbad466
Wang T, Sun J, Zhao Q (2023) Investigating cardiotoxicity related with hERG channel blockers using molecular fingerprints and graph attention mechanism. Comput Biol Med 153:106464. https://doi.org/10.1016/j.compbiomed.2022.106464
Wei D, Peslherbe GH, Selvaraj G et al (2022) Advances in drug design and development for human therapeutics using artificial intelligence—I. Biomolecules 12(12):1846. https://doi.org/10.3390/biom12121846
Peng J, Hui W, Li Q et al (2019) A learning-based framework for miRNA–disease association identification using neural networks. Bioinformatics 35(21):4364–4371. https://doi.org/10.1093/bioinformatics/btz254
Chu Y, Zhang Y, Wang Q et al (2022) A transformer-based model to predict peptide-HLA class I binding and optimize mutated peptides for vaccine design. Nat Mach Intell 4(3):300–311. https://doi.org/10.1038/s42256-022-00459-7
Chen Y, Wang J, Wang C et al (2022) Deep learning models for disease-associated circRNA prediction: a review. Brief Bioinform 23(6):bbac364. https://doi.org/10.1093/bib/bbac364
Sun F, Sun J, Zhao Q (2022) A deep learning method for predicting metabolite–disease associations via graph neural network. Brief Bioinform 23(4):bbac266. https://doi.org/10.1093/bib/bbac266
Lin S, Chen W, Chen G et al (2022) MDDI-SCL: predicting multi-type drug–drug interactions via supervised contrastive learning. J Cheminform 14(1):1–12. https://doi.org/10.1186/s13321-022-00659-8
Peng L, Yuan R, Han C et al (2023) CellEnBoost: a boosting-based ligand–receptor interaction identification model for cell-to-cell communication inference. IEEE Trans Nanobiosci 22(4):705–715. https://doi.org/10.1109/TNB.2023.3278685
Xu J, Xu J, Meng Y et al (2023) Graph embedding and Gaussian mixture variational autoencoder network for end-to-end analysis of single-cell RNA sequencing data. Cell Rep Methods 3(1):100382. https://doi.org/10.1016/j.crmeth.2022.100382
Peng L, He X, Peng X et al (2023) STGNNks: identifying cell types in spatial transcriptomics data based on graph neural network, denoising auto-encoder, and k-sums clustering. Comput Biol Med 166:107440. https://doi.org/10.1016/j.compbiomed.2023.107440
Zhou Z, Zhuo L, Fu X et al (2024) Joint masking and self-supervised strategies for inferring small molecule–miRNA associations. Mol Ther-Nucl Acids 35(1):102103. https://doi.org/10.1016/j.omtn.2023.102103
Peng L, Xiong W, Han C et al (2024) Cell dialog: a computational framework for ligand–receptor-mediated cell–cell communication analysis. IEEE J Biomed Health 28(1):580–591. https://doi.org/10.1109/JBHI.2023.3333828
Yang Q, Li X (2021) BiGAN: LncRNA–disease association prediction based on bidirectional generative adversarial network. BMC Bioinform 22:1–17. https://doi.org/10.1186/s12859-021-04273-7
Wang W, Dai Q, Li F et al (2021) MLCDForest: multi-label classification with deep forest in disease prediction for long non-coding RNAs. Brief Bioinform 22(3):bbaa104. https://doi.org/10.1093/bib/bbaa104
Sheng N, Cui H, Zhang T et al (2021) Attentional multi-level representation encoding based on convolutional and variance autoencoders for lncRNA–disease association prediction. Brief Bioinform 22(3):bbaa067. https://doi.org/10.1093/bib/bbaa067
Sheng N, Wang Y, Huang L et al (2023) Multi-task prediction-based graph contrastive learning for inferring the relationship among lncRNAs, miRNAs and diseases. Brief Bioinform 24(5):bbad276. https://doi.org/10.1093/bib/bbad276
Fan Y, Chen M, Pan X (2022) GCRFLDA: scoring lncRNA–disease associations using graph convolution matrix completion with conditional random field. Brief Bioinform 23(1):bbab361. https://doi.org/10.1093/bib/bbab361
Ma Y (2022) DeepMNE: deep multi-network embedding for lncRNA–disease association prediction. IEEE J Biomed Health 26(7):3539–3549. https://doi.org/10.1109/JBHI.2022.3152619
Zhu Y, Zhang F, Zhang S et al (2023) Predicting latent lncRNA and cancer metastatic event associations via variational graph auto-encoder. Methods 211:1–9. https://doi.org/10.1016/j.ymeth.2023.01.006
Zhao X, Zhao X, Yin M (2022) Heterogeneous graph attention network based on meta-paths for lncRNA–disease association prediction. Brief Bioinform 23(1):bbab407. https://doi.org/10.1093/bib/bbab407
Zhou Y, Wang X, Yao L et al (2022) LDAformer: predicting lncRNA–disease associations based on topological feature extraction and transformer encoder. Brief Bioinform 23(6):bbac370. https://doi.org/10.1093/bib/bbac370
Xuan P, Gong Z, Cui H et al (2022) Fully connected autoencoder and convolutional neural network with attention-based method for inferring disease-related lncRNAs. Brief Bioinform 23(3):bbac089. https://doi.org/10.1093/bib/bbac089
Liu Y, Yu Y, Zhao S (2022) Dual attention mechanisms and feature fusion networks based method for predicting LncRNA–disease associations. Interdiscip Sci 14(2):358–371. https://doi.org/10.1007/s12539-021-00492-x
Xuan P, Bai H, Cui H et al (2023) Specific topology and topological connection sensitivity enhanced graph learning for lncRNA–disease association prediction. Comput Biol Med 164:107265. https://doi.org/10.1016/j.compbiomed.2023.107265
Chen G, Wang Z, Wang D et al (2012) LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 41(D1):D983–D986. https://doi.org/10.1093/nar/gks1099
Cui T, Zhang L, Huang Y et al (2018) MNDR v2.0: an updated resource of ncRNA–disease associations in mammals. Nucleic Acids Res 46(D1):D371–D374. https://doi.org/10.1093/nar/gkx1025
Lowe HJ, Barnett GO (1994) Understanding and using the medical subject headings (MeSH) vocabulary to perform literature searches. Jama 271(14):1103–1108. https://doi.org/10.1001/jama.1994.03510380059038
Chen X, Clarence Yan C, Luo C et al (2015) Constructing lncRNA functional similarity network based on lncRNA–disease associations and disease semantic similarity. Sci Rep-UK 5(1):11338. https://doi.org/10.1038/srep11338
Wang L, Wong L, Li Z et al (2022) A machine learning framework based on multi-source feature fusion for circRNA–disease association prediction. Brief Bioinform 23(5):bbac388. https://doi.org/10.1093/bib/bbac388
Fan W, Shang J, Li F et al (2020) IDSSIM: an lncRNA functional similarity calculation model based on an improved disease semantic similarity method. BMC Bioinform 21(1):1–14. https://doi.org/10.1186/s12859-020-03699-9
Chen X, Huang Y-A, Wang X-S et al (2016) FMLNCSIM: fuzzy measure-based lncRNA functional similarity calculation model. Oncotarget 7(29):45948. https://doi.org/10.18632/oncotarget.10008
Wang D, Wang J, Lu M et al (2010) Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics 26(13):1644–1650. https://doi.org/10.1093/bioinformatics/btq241
Wang Y-X, Zhang Y (2012) Nonnegative matrix factorization: a comprehensive review. IEEE Trans Knowl Data Eng 25(6):1336–1353. https://doi.org/10.1109/TKDE.2012.51
Ding Y, Lei X, Liao B et al (2021) Predicting miRNA–disease associations based on multi-view variational graph auto-encoder with matrix factorization. IEEE J Biomed Health 26(1):446–457. https://doi.org/10.1109/JBHI.2021.3088342
Salehi A, Davulcu H (2019) Graph attention auto-encoders. arXiv. https://doi.org/10.48550/arXiv.1905.10715
Zhou Z, Zhuo L, Fu X et al (2024) Joint deep autoencoder and subgraph augmentation for inferring microbial responses to drugs. Brief Bioinform 25(1):bbad483. https://doi.org/10.1093/bib/bbad483
Peng L, Huang L, Tian G et al (2023) Predicting potential microbe–disease associations with graph attention autoencoder, positive-unlabeled learning, and deep neural network. Front Microbiol 14:1244527. https://doi.org/10.3389/fmicb.2023.1244527
Veliçković P, Cucurull G, Casanova A et al (2017) Graph attention networks. arXiv. https://doi.org/10.48550/arXiv.1710.10903
Deng L, Liu Z, Qian Y et al (2022) Predicting circRNA–drug sensitivity associations via graph attention auto-encoder. BMC Bioinform 23(1):1–15. https://doi.org/10.1186/s12859-022-04694-y
Wu X, Lu W, Quan Y et al (2024) Deep dual graph attention auto-encoder for community detection. Expert Syst Appl 238:122182. https://doi.org/10.1016/j.eswa.2023.122182
Wojtas M, Chen K (2020) Feature importance ranking for deep learning. Adv Neural Inf Process Syst 33:5105–5114. https://proceedings.neurips.cc/paper_files/paper/2020/file/36ac8e558ac7690b6f44e2cb5ef93322-Paper.pdf
Lihong P, Wang C, Tian X et al (2022) Finding lncRNA–protein interactions based on deep learning with dual-net neural architecture. IEEE ACM Trans Comput Biol 19(6):3456–3468. https://doi.org/10.1109/TCBB.2021.3116232
Mengshoel OJ (2008) Understanding the role of noise in stochastic local search: analysis and experiments. Artif Intell 172(8–9):955–990. https://doi.org/10.1016/j.artint.2007.09.010
Soydaner D (2020) A comparison of optimization algorithms for deep learning. Int J Pattern Recognit 34(13):2052013. https://doi.org/10.1142/S0218001420520138
Hailesilassie T (2016) Rule extraction algorithm for deep neural networks: a review. arXiv. https://doi.org/10.48550/arXiv.1610.05267
Ding L, Wang M, Sun D et al (2018) TPGLDA: novel prediction of associations between lncRNAs and diseases via lncRNA-disease-gene tripartite graph. Sci Rep-UK 8(1):1065. https://doi.org/10.1038/s41598-018-19357-3
Guo Z, You Z, Wang Y-B et al (2019) A learning-based method for LncRNA–disease association identification combing similarity information and rotation forest. IScience 19:786–795. https://doi.org/10.1016/j.isci.2019.08.030
Zeng M, Lu C, Zhang F et al (2020) SDLDA: lncRNA–disease association prediction based on singular value decomposition and deep learning. Methods 179:73–80. https://doi.org/10.1016/j.ymeth.2020.05.002
Zhang Y, Ye F, Xiong D et al (2020) LDNFSGB: prediction of long non-coding RNA and disease association using network feature similarity and gradient boosting. BMC Bioinform 21(1):1–27. https://doi.org/10.1186/s12859-020-03721-0
Kumar S, Sharma AK, Lalhlenmawia H et al (2021) Natural compounds targeting major signaling pathways in lung cancer. In: Dua K, Löbenberg R, Malheiros Luzo ÂC, Shukla S, Satija S (eds) Targeting cellular signalling pathways in lung diseases. Springer, Singapore, pp 821–846. https://doi.org/10.1007/978-981-33-6827-9_37
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Ismail A, El-Mahdy HA, Abulsoud AI et al (2023) Beneficial and detrimental aspects of miRNAs as chief players in breast cancer: a comprehensive review. Int J Biol Macromol 224:1541–1565. https://doi.org/10.1016/j.ijbiomac.2022.10.241
Wilkinson L, Gathani T (2022) Understanding breast cancer as a global health concern. Brit J Radiol 95(1130):20211033. https://doi.org/10.1259/bjr.20211033
Arnold M, Morgan E, Rumgay H et al (2022) Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast 66:15–23. https://doi.org/10.1016/j.breast.2022.08.010
Chen J, Lin J, Hu Y et al (2023) RNADisease v4.0: an updated resource of RNA-associated diseases, providing RNA-disease analysis, enrichment and prediction. Nucleic Acids Res 51(D1):D1397–D1404. https://doi.org/10.1093/nar/gkac814
Gao Y, Shang S, Guo S et al (2021) Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res 49(D1):D1251–D1258. https://doi.org/10.1093/nar/gkaa1006
Bao Z, Yang Z, Huang Z et al (2019) LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res 47(D1):D1034–D1037. https://doi.org/10.1093/nar/gky905
Aliquò F, Minuti A, Avenoso A et al (2023) Endocan promotes pro-tumorigenic signaling in lung cancer cells: modulation of cell proliferation, migration and lncRNAs H19 and HULC expression. Int J Mol Sci 24(9):8178. https://doi.org/10.3390/ijms24098178
Xu Y, Li J, Wang P et al (2019) LncRNA HULC promotes lung squamous cell carcinoma by regulating PTPRO via NF-\(\kappa \)B. J Cell Biochem 120(12):19415–19421. https://doi.org/10.1002/jcb.29119
Padua D, Mahurkar-Joshi S, Law IKM et al (2016) A long noncoding RNA signature for ulcerative colitis identifies IFNG-AS1 as an enhancer of inflammation. Am J Physiol Gastrointest Liver Physiol 311(3):G446–G457. https://doi.org/10.1152/ajpgi.00212.2016
Gomez JA, Wapinski OL, Yang YW et al (2013) The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-\(\gamma \) locus. Cell 152(4):743–754. https://doi.org/10.1016/j.cell.2013.01.015
Ye J, Chen X, Lu W (2021) Identification and experimental validation of immune-associate lncRNAs for predicting prognosis in cervical cancer. Oncotargets Ther 14:4721. https://doi.org/10.2147/OTT.S322998
Zhu L-L, Wu Z, Li R-K et al (2021) Deciphering the genomic and lncRNA landscapes of aerobic glycolysis identifies potential therapeutic targets in pancreatic cancer. Int J Biol Sci 17(1):107. https://doi.org/10.7150/ijbs.49243
Zhou L, Li H, Sun T et al (2022) HULC targets the IGF1R–PI3K-AKT axis in trans to promote breast cancer metastasis and cisplatin resistance. Cancer Lett 548:215861. https://doi.org/10.1016/j.canlet.2022.215861
Leisegang MS, Bains JK, Seredinski S et al (2022) HIF1\(\alpha \)-AS1 is a DNA:DNA:RNA triplex-forming lncRNA interacting with the HUSH complex. Nat Commun 13(1):6563. https://doi.org/10.1038/s41467-022-34252-2
Dalmasso B, Ghiorzo P (2023) Long non-coding RNAs and metabolic rewiring in pancreatic cancer. Cancers 15(13):3486. https://doi.org/10.3390/cancers15133486. https://www.mdpi.com/2072-6694/15/13/3486
Zhang H, Wang X, Wu Y et al (2021) Deciphering the molecular mechanism of long non-coding RNA HIFA-AS1 regulating pancreatic cancer cells. Res Square. https://doi.org/10.21203/rs.3.rs-533849/v1
Cao M, Luo H, Li D et al (2022) Research advances on circulating long noncoding RNAs as biomarkers of cardiovascular diseases. Int J Cardiol 353:109–117. https://doi.org/10.1016/j.ijcard.2022.01.070
Yan Y, Wu D, Yang L et al (2023) The expanding roles of long non-coding RNAs in pancreatic cancer. Adv Ther 6(12):202300178. https://doi.org/10.1002/adtp.202300178
Wang Z, Cao Z, Wang Z (2021) Significance of long non-coding RNA IFNG-AS1 in the progression and clinical prognosis in colon adenocarcinoma. Bioengineered 12(2):11342–11350. https://doi.org/10.1080/21655979.2021.2003944
Xu F, Huang M, Chen Q et al (2021) LncRNA HIF1A-AS1 promotes gemcitabine resistance of pancreatic cancer by enhancing glycolysis through modulating the AKT/YB1/HIF1\(\alpha \) pathway. Cancer Res 81(22):5678–5691. https://doi.org/10.1158/0008-5472.CAN-21-0281
Acknowledgements
We are very thankful for three anonymous reviewers and all authors of references.
Funding
L.H. Peng was supported by National Natural Science Foundation of China under Grant nos. 62072172 and 61803151 and Natural Science Foundation of Hunan Province of China under Grant 2023JJ50201. M. Chen was supported by National Natural Science Foundation of China under Grant no. 62172158.
Author information
Authors and Affiliations
Contributions
L.H. Peng and L.L. Huang: conceptualization; L.H. Peng, and M. Chen: funding acquisition; L.H. Peng, L.L. Huang, and M. Chen: project administration; L.H. Peng and M.N. Ren: writing—original draft; L.H. Peng and M. Chen: writing—review and editing; L.H. Peng, M.N. Ren, L.L. Huang, and M. Chen: investigation; L.H. Peng, M.N. Ren, and L.L. Huang: methodology; L.L. Huang and M.N. Ren: software; L.H. Peng, L.L. Huang, and M. Chen: validation. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peng, L., Ren, M., Huang, L. et al. GEnDDn: An lncRNA–Disease Association Identification Framework Based on Dual-Net Neural Architecture and Deep Neural Network. Interdiscip Sci Comput Life Sci (2024). https://doi.org/10.1007/s12539-024-00619-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12539-024-00619-w